INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) is one of the major causes of diarrhoea in children and travellers in developing countries [Reference Qadri1, Reference Clarke2]. The ability of ETEC to adhere to and colonize the intestinal epithelium is an essential step for pathogenicity in addition to its ability to produce heat-labile enterotoxin (LT) and/or heat-stable enterotoxin (ST). The colonizing ability of human ETEC depends on the presence of colonization factors (CFs) on the surface of the cells, which usually form pili, also known as fimbriae [Reference Kaper, Nataro and Mobley3, Reference Nataro and Kaper4]. Several types of colonization factor antigens (CFAs) and putative colonization factors (PCFs) have been identified on the basis of antigenic specificity and/or N-terminal amino-acid sequence of the major subunit (pilin), e.g. CFA/I, CFA/II, CS8 (originally CFA/III), CFA/IV, CS12 (PCFO159), CS13 (PCFO9), CS14 (PCFO166), CS15 (antigen 8786), CS17, CS18 (PCFO20), CS19 and CS20 [Reference Torres, Zhou and Kaper5, Reference Gaastra and Svennerholm6]. Of these, CFA/II and CFA/IV are heterogeneous and consist of a complex of different antigens named coli surface (CS) antigens. CFA/II is composed of CS1, CS2 and CS3, which are present in different permutations. Similarly, CFA/IV is composed of CS4, CS5 and CS6. CFA/II-producing ETEC strains express CS3 alone or in combination with CS1 or CS2, while CFA/IV-producing ETEC strains express CS6 alone or together with CS4 or CS5. Although at least over 20 different CFs are known in human ETEC, there is still a substantial proportion of strains on which CFs have not been identified, and additional CFs may thus exist.
CFAs have been proposed as candidates for inclusion in a pilus vaccine against ETEC diarrhoea [Reference Walker, Steele and Aguado7, Reference Girard8]. In order to formulate such a vaccine, it is important to know the distribution of CFs on ETEC strains in different areas of the world. The identification and typing of CF is determined by serological test with specific monoclonal and/or polyclonal antibodies against each CF. However, such identification and typing method can be used in only a limited number of reference laboratories [Reference Jiang9–Reference Al-Gallas11]. We therefore designed polymerase chain reaction (PCR) primers specific for each CF for the development of a simple PCR-based genotypic CF identification method. This method was applied to 17 randomly selected clinical isolates of ETEC strains in Thailand.
MATERIALS AND METHODS
Bacterial strains
A total of 17 ETEC strains were originally isolated from stool samples obtained from different patients with diarrhoea in Thailand between May and September 2006. All strains were serogrouped with commercially available antisera (Denka Seiken Co. Ltd, Japan) for specific somatic (O) antigens by an established method, and stored in Luria–Bertani (LB) broth [Reference Sambrook and Russell12] containing 25% glycerol at −80°C at the Department of Medical Sciences, Ministry of Public Health, Thailand. Immediately before CF analyses, all strains were rechecked for enterotoxin (LT, STh and STp) genes by PCR, since it is well known that enterotoxin genes are usually co-localized with CF genes on virulence plasmids. Strains that did not possess any enterotoxin genes at the time of these analyses were therefore assumed to have lost their CF genes and omitted from this study.
Bacterial culture conditions
ETEC strains were routinely grown on heart infusion agar plates (Difco, USA) or in LB broth at 37°C for 20 h [Reference Sambrook and Russell12]. For optimal expression of CF, ETEC strains were grown on CFA agar plates at 37°C for 20 h [Reference Evans, Evans and Tjoa13].
Primer design
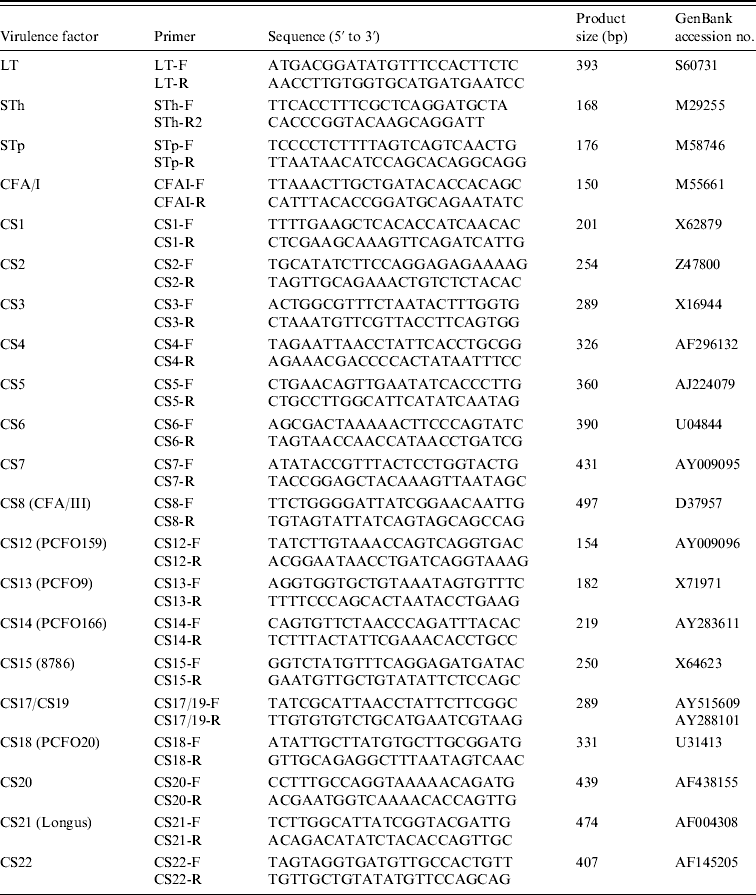
Nucleotide sequences of different major pilins were aligned to identify unique regions and utilized to design the CF-specific PCR primers (Table 1), except for the CS17/19-F and CS17/19-R primers that might recognize both CS17 and CS19 genes, since these nucleotide sequences are very similar. The specificity of the primers was tested by BLAST search.
Table 1. PCR primers used in this study
DNA extraction and PCR
An overnight broth culture (10 μl) was added to 90 μl of distilled water, boiled for 5 min and centrifuged at 12 000 g for 10 min at 4°C. The supernatant was used as the DNA template. PCR was performed with a 50-μl reaction mixture containing 1× PCR buffer (Takara Bio Inc., Japan), dNTP mixture (2·5 mm each of dATP, dTTP, dCTP and dGTP; Takara Bio Inc.), 0·5 μm of each primer, 10 μl of DNA template and 1·25 U of TaKaRa Ex TaqTM (Takara Bio Inc.), and the mixture was then subjected to PCR amplification using a PCR Thermal Cycler (Gene Amp PCR system 9700; Applied Biosystems, USA) under the following conditions: 95°C for 1 min, 52°C for 1 min and 72°C for 1 min for 35 cycles, with a final extension at 72°C for 5 min. PCR products were subsequently subjected to 2% agarose gel electrophoresis, stained with ethidium bromide and photographed under ultraviolet light.
Salting-out test (hydrophobicity test)
The salting-out test was performed as described by Honda et al. [Reference Honda14]. Bacterial hydrophobicity was determined by observing cell clumping in ammonium sulphate solutions at a range of concentrations (0·25, 0·5, 1·0, 2·0 and 4·0 m). The lower the concentration of ammonium sulphate, the higher the cell-surface hydrophobicity is.
Caco-2 adhesion test
ETEC strains were subjected to a Caco-2 adhesion test with the method described by Taniguchi et al. [Reference Taniguchi15]. The adhesion indices were presented as the percentage of Caco-2 cells with at least one adhering bacterium (index 1) and the average number of bacteria/cell (index 2) by counting 10 randomly chosen fields in three separate experiments.
RESULTS AND DISCUSSION
This study focused on the examination of 17 randomly selected clinical isolates of ETEC in a collection of E. coli strains kept at the Department of Medical Sciences, Ministry of Public Health, Thailand. The characteristics of these 17 ETEC strains are summarized in Table 2.
Table 2. Characteristics of ETEC strains isolated from patients with diarrhoea in Thailand

* Not agglutinated in 4·0 m ammonium sulphate solution.
† PCR cannot distinguish between CS17 and CS19.
PCR of toxin genes
The ETEC strains were rechecked for enterotoxin (LT, STh, STp) genes by PCR, which resulted in the detection of the genes encoding LT (eight strains, 47%) or STp (five strains, 29%) or STh (4 strains, 24%). No positive strain was detected which possessed genes encoding both LT and ST (STh or STp) (Table 2).
PCR of CF genes
Figure 1 shows the results of agarose gel electrophoresis of PCR-amplified products for detection of the CS6 gene. ETEC EC859/49, ETEC EC860/49, ETEC EC861/49, ETEC EMEC 134/49 and EMEC 140-1/49 yielded specific PCR-amplified products with the expected size (390 bp) for the CS6 gene (Fig. 1, lanes 6–8, 16, 17). Since some non-specific PCR-amplified products were observed in the lanes of CS6-negative strains, we attempted to optimize the PCR conditions for the examination of several important parameters such as annealing temperature, primer concentrations and magnesium ion concentration. Unfortunately, the findings did not improve significantly. Of the 17 ETEC strains, five were CS6-gene positive (29%), followed by CS13-gene positive (12%), CFA/I-, CS2 and CS3- and CS17/CS19-gene positive (6% each), while seven strains (41%) were negative for other CF genes (Table 2). Four of the five CS6-gene positive strains, each isolated from a different patient, were O159 and STp-gene positive. We intend to analyse these strains in our laboratory for any epidemiological clonal relationships. Two CS13 and LTh-gene positive strains will also be investigated.
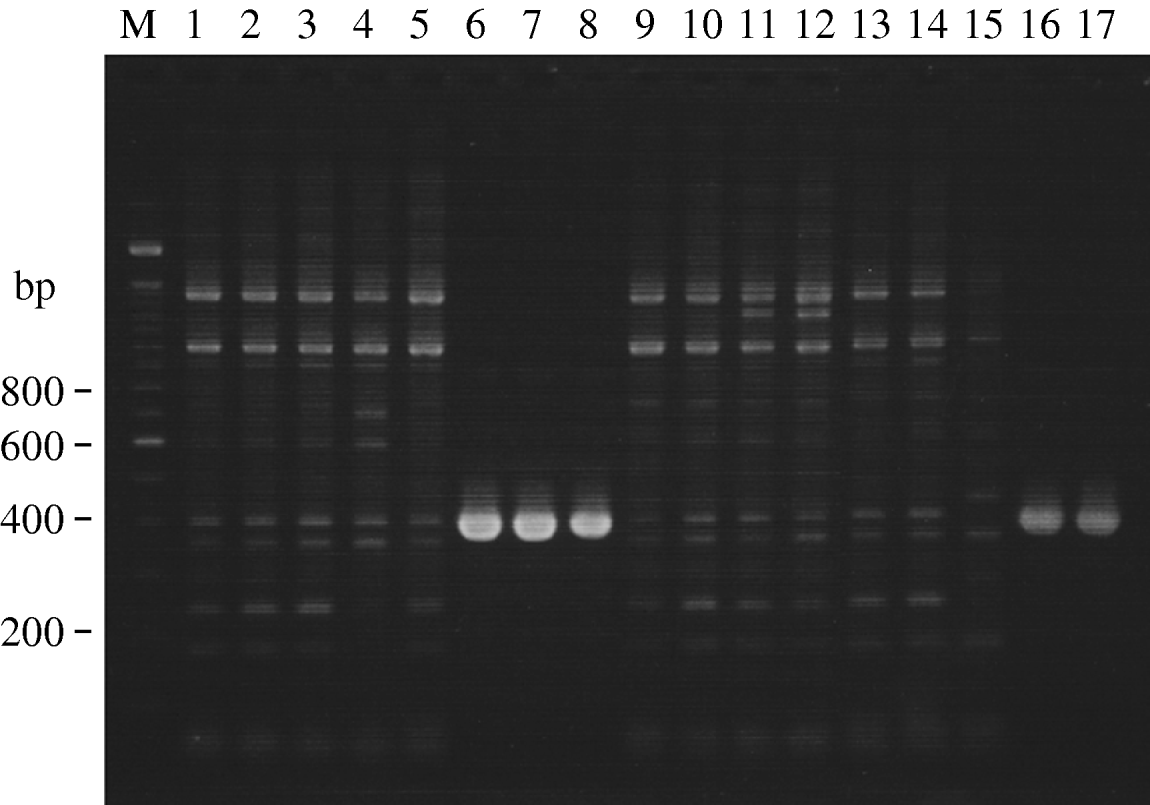
Fig. 1. Agarose gel electrophoresis showing PCR-amplified products for detection of CS6 gene in clinical isolates of ETEC strains. CS6-gene primers CS6-F and CS6-R were used. Lane M, 100-bp DNA ladder marker (Invitrogen, USA); lanes 1–17, clinical isolates of ETEC strains correspond to those listed in Table 2.
Previous surveillance of ETEC strains in different areas of the world showed wide variations in CFs [Reference Sommerfelt16, Reference Nirdnoy17]. In the present study, diarrhoea due to CS6-producing ETEC is now recognized as one of the most common and important ETEC infections (25–35%) in the world [Reference Jiang9–Reference Al-Gallas11]. Our data for the strains in Thailand, obtained between May and September 2006, are consistent with these findings in that 29% of the cases may be due to this phenotype.
Salting-out test (hydrophobicity test)
Of the 17 ETEC strains, seven (41%) agglutinated at a relatively low concentration (<0·5 m) of ammonium sulphate, indicating a high degree of cell-surface hydrophobicity (Table 2). CFA/I-, CS2-, CS13- and CS17/CS19-gene positive strains corresponded to highly hydrophobic cells, which are morphologically pilus structures known as rigid rod-shaped pili or flexible pili, whereas the two hydrophobic strains (EMEC 107-2/49 and EMEC 155/49) could not amplify CF genes as determined by PCR. None of the CS6-gene positive strains agglutinated in 4·0 m ammonium sulphate, indicating a lower degree of cell-surface hydrophobicity, which is consistent with the fact that CS6 is known as a non-pilus structure [Reference Lüdi18].
Caco-2 adhesion test
ETEC strains were tested for the ability to adhere to the Caco-2 cells, an established cell culture model for ETEC colonization. For example, the ability of ETEC EC18/49 (CF-gene negative strain), ETEC EC60/49 (CS13-gene positive strain), ETEC EC428/49 (CFA/I-gene positive strain) and ETEC EC859/49 (CS6-gene positive strain) to adhere to the Caco-2 cells is demonstrated in Figure 2. The ETEC strains adhered to the Caco-2 cells with indices (index 1) of 89·8, 31·3, 72·8 and 84·3%, respectively, and with an average number of bacteria/cell (index 2) of 86·8, 0·8, 18·6 and 48·4, respectively. Adherence indices of CS6-gene positive strains were higher than those of other CF-gene positive strains (Table 2). Interestingly, the Caco-2 adhesion test also revealed that ETEC EC18/49 was negative for any CF genes and non-hydrophobic strains in the salting-out test, but adhered to the Caco-2 cells, suggesting that ETEC EC18/49 may posses an as yet unknown adherence factor. We intend to investigate this further.
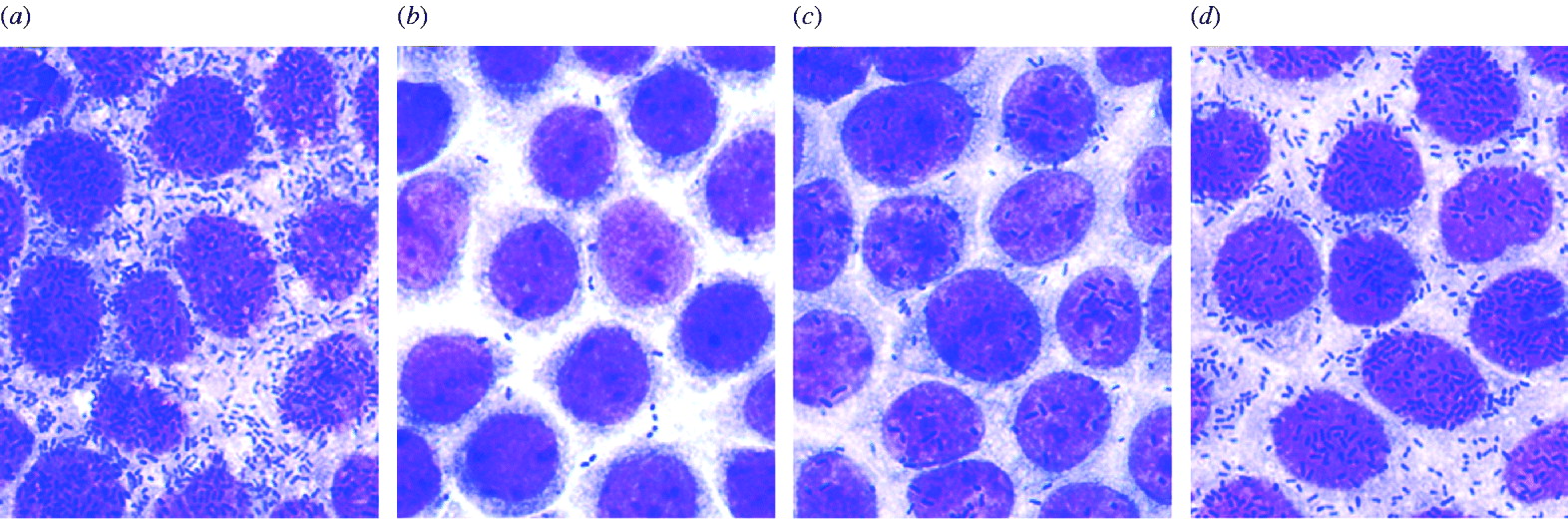
Fig. 2. Micrographs showing adhesion of ETEC strains to Caco-2 cells. (a) ETEC EC18/49 (CF-gene negative strain); (b) ETEC EC60/49 (CS13-gene positive strain); (c) ETEC EC428/49 (CFA/I-gene positive strain); (d) ETEC EC859/49 (CS6-gene positive strain).
Several phenotypic and genotypic identification methods have been developed for the identification of CFs [Reference Jiang9–Reference Al-Gallas11, Reference Sommerfelt16, Reference Nirdnoy17]. The bacterial agglutination assay using anti-CF antibodies can be performed quickly, but it requires specific antibodies. Genetic identification methods such as DNA hybridization can also be used, but they require enzyme-labelling of the probes. Recently, and independently from our study, Rodas et al. described the development of multiplex PCR assays for identification of 19 ETEC CFs, and concluded that the PCR-based identification method was superior to the phenotypic colony dot-blot assay using anti-CF antibodies, since false-negative results were found in the latter. [Reference Rodas19]. In view of their report and our present study, the PCR assay is a simple and effective method with several advantages compared to other methods for CF detection, we therefore recommend this system for elucidating the epidemiology of ETEC infection, and for aiding the development of an ideal CF vaccine against ETEC diarrhoea.
In summary, CS6-producing ETEC is the most prevalent strain of ETEC in Thailand, which is similar to the situation reported in various developing countries [Reference Jiang9–Reference Al-Gallas11]. Most CS6-producing ETEC strains express ST (LT/ST or only ST), but immunity to ST is difficult to stimulate. CS6 should therefore be considered as a target for any ETEC vaccine development.
ACKNOWLEDGEMENTS
We thank Dr Orn-Anong Ratchtrachenchai and Ms. Sasitorn Rakyart, National Institute of Health for providing clinical isolates of ETEC strains and for their assistance. We are also grateful to Dr Yoshitake Nishimune from the Research Collaboration Center for Emerging and Re-emerging Infections (RCC-ERI) for his valuable help. This work was supported in part by the programme of the RCC-ERI launched by a project commissioned by the Ministry of Education, Cultures, Sports, Science and Technology of Japan in collaboration with the Department of Medical Sciences of the Ministry of Public Health, Thailand.
DECLARATION OF INTEREST
None.








