No CrossRef data available.
Article contents
On a wider class of prior distributions for graphical models
Published online by Cambridge University Press: 08 June 2023
Abstract
Gaussian graphical models are useful tools for conditional independence structure inference of multivariate random variables. Unfortunately, Bayesian inference of latent graph structures is challenging due to exponential growth of 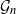 $\mathcal{G}_n$, the set of all graphs in n vertices. One approach that has been proposed to tackle this problem is to limit search to subsets of
$\mathcal{G}_n$, the set of all graphs in n vertices. One approach that has been proposed to tackle this problem is to limit search to subsets of 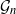 $\mathcal{G}_n$. In this paper we study subsets that are vector subspaces with the cycle space
$\mathcal{G}_n$. In this paper we study subsets that are vector subspaces with the cycle space 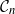 $\mathcal{C}_n$ as the main example. We propose a novel prior on
$\mathcal{C}_n$ as the main example. We propose a novel prior on 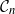 $\mathcal{C}_n$ based on linear combinations of cycle basis elements and present its theoretical properties. Using this prior, we implement a Markov chain Monte Carlo algorithm, and show that (i) posterior edge inclusion estimates computed with our technique are comparable to estimates from the standard technique despite searching a smaller graph space, and (ii) the vector space perspective enables straightforward implementation of MCMC algorithms.
$\mathcal{C}_n$ based on linear combinations of cycle basis elements and present its theoretical properties. Using this prior, we implement a Markov chain Monte Carlo algorithm, and show that (i) posterior edge inclusion estimates computed with our technique are comparable to estimates from the standard technique despite searching a smaller graph space, and (ii) the vector space perspective enables straightforward implementation of MCMC algorithms.
Keywords
MSC classification
- Type
- Original Article
- Information
- Copyright
- © The Author(s), 2023. Published by Cambridge University Press on behalf of Applied Probability Trust





