Human milk is often the sole dietary source for the first few months of life. It contains not only all the nutrients necessary for infants to thrive, but also ingredients that may provide health benefits beyond those of traditional nutrients. Human milk oligosaccharides (HMO) are part of these functional ingredients. Given the vast quantities of oligosaccharides that are present in human milk (concentrations reaching up to 50 g/l or more in the colostrum to an average of 10–15 g/l in mature milk(Reference Kunz, Rodriguez-Palmero and Koletzko1, Reference Kuntz, Rudloff and Kunz2)), it is hardly surprising that during postnatal development, these molecules play an important physiological role. Research on HMO and glycoconjugates has received much attention in recent years, and there is increasing evidence of the local effects of HMO within the gastrointestinal tract. Such effects may include prebiotic, anti-adhesive and anti-inflammatory activities, glycome modification, an influence on brain development and growth-related characteristics of intestinal cells and other uncharacterised effects(Reference Kuntz, Rudloff and Kunz2, Reference Newburg, Ruiz-Palacios and Morrow3–Reference Hickey and Corredig5).
There is accumulating evidence that milk oligosaccharides also have direct effects on the maturation of the immune system. At birth, although developed, the lymphoid system is not yet mature and the immune system is dominated by helper T-cells (Th) of subtype-2. Maturation of the immune system subsequently occurs and Th of subtype-1 become the more dominant cell type(Reference Field, Caccavelli and Fillion6, Reference Schack-Nielsen and Michaelsen7). Th1 responses are pro-inflammatory and high levels are associated with Crohn's and autoimmune diseases, while Th2 responses are immunoregulatory and high levels are associated with asthma and atrophic dermatitis(Reference Romagnani8). Therefore, striking the right balance with regard to lymphoid cell development is a complex process involving the timely expression of growth factors (cytokines and chemokines), receptors and adhesion molecules(Reference Niers, Hoekstra and Timmerman9). It is now apparent that the oligosaccharide component of human milk may play a role in the modulation of the T-cell type. For example, Velupillai & Harn(Reference Velupillai and Harn10) demonstrated that neutral HMO, lacto-N-fucopentaose III and lacto-N-neotetraose, stimulated B-cells to proliferate and induce the production of immunomodulatory cytokines such as IL-10, which is known to down-regulate Th1 cells. A further study by Eiwegger et al. (Reference Eiwegger, Stahl and Schmitt11) has shown that acidic HMO had an immunomodulatory effect on stimulated cord blood T-cells in vitro. In their study, acidic HMO altered the cytokine profile of allergy-specific T-cells, resulting in a more balanced Th1/Th2 profile in cord blood cells. Similarly, Vos et al. (Reference Vos, Haarman and van Ginkel12) demonstrated that acidic milk oligosaccharides induced Th1-skewing effects in a murine model. The above-mentioned studies have provided evidence that specific HMO may have a direct role in the postnatal maturation of the immune system and may result in allergy prevention in breast-fed infants once absorbed. However, before investigating the activity of such molecules at the systemic level, a full understanding of what is happening at the surface of the epithelial cells is required, as these are the first cell type that the oligosaccharides come in contact with: for example, understanding the effect of milk oligosaccharides on the production of chemokines and cytokines, and the expression of cell surface receptors, by gut epithelial cells.
Since human milk is not readily available as a commercial feedstock, the food industry must focus its attention on alternative, sustainable sources of bioactive oligosaccharides. Bovine milk is a candidate source of oligosaccharides that may mimic the biological health benefits of HMO. Indeed, it has been shown to contain complex oligosaccharides that share a high degree of structural similarity to those of human milk(Reference Tao, DePeters and Freeman13–Reference Marino, Lane and Abrahams16). Furthermore, some studies have suggested that bovine milk oligosaccharides (BMO) have biological effects similar to those of HMO. For example, BMO have been shown to reduce the adherence of pathogens, such as Neisseria meningitidis, Helicobacter pylori, Campylobacter jejuni and influenza virus, to their respective ligands(Reference Mysore, Wigginton and Simon17–Reference Lane, Marino and Naughton20). Whether BMO have similar effects on the immune system when compared with HMO remains unknown. However, Otani & Monnai(Reference Otani and Monnai21) found that the sialic acid fraction of bovine glycomacropeptide may modulate the immune system in a mouse model.
To gain insight into the comparative transcriptional responses of the gut epithelium to milk oligosaccharides, we compared the transcriptomes of HT-29 colonic epithelial cells after exposure to either the entire pool of HMO or bovine colostrum oligosaccharides (BCO). Specifically, we investigated their contribution to the development of the intestinal immune response by concentrating on the expression profiles of immune system-associated glycogenes.
Materials and methods
Isolation of milk oligosaccharides
Free oligosaccharides were isolated from bovine and human colostrum (day 1) obtained from Holstein Friesian cattle on-site at the Teagasc Food Research Centre, Moorepark (Fermoy, Cork, Ireland), and the Irvinestown Human Milk Bank (County Fermanagh, Ireland), respectively. Both samples were defatted and deproteinised through conventional methods as described by Kobata & Ginsburg(Reference Kobata and Ginsburg22). In brief, aliquots were centrifuged at 5000 rpm for 20 min at 4°C in a Sorvall RC6 plus® to separate the fat. The aqueous phase was collected and 1 m-HCl was added until a pH of 4·6 was reached. The sample was then heated to 35°C for 2 h and precipitated caseins were removed by centrifugation at 5000 rpm for 30 min at 25°C. The pH of the supernatant was then neutralised using 4 m-NaOH and the supernatant was ultrafiltered through a 5 kDa molecular weight cut-off membrane (Millipore® Helicon S10 spiral cartridge; Millapore) to remove large peptides. Oligosaccharides present in the permeate were collected, freeze-dried and stored at − 80°C. To remove residual peptides and deplete the high levels of lactose, a 20 % solution (40 ml) of the freeze-dried powder was applied to a Sephadex G-25 column (92 × 2·6 cm; Pharmacia) and eluted with deionised water at a flow rate of 5 ml/min. The fractions were monitored for peptides(Reference Bradford23), lactose and sialyllactose using high-pH anion-exchange chromatography with pulsed amperometric detection. Peptide-free fractions with little or no lactose were pooled to form a pool of purified oligosaccharides. The oligosaccharide standard 3′-sialyllactose was purchased from Carbosynth Limited.
Cell culture and oligosaccharide exposure
The human colon adenocarcinoma cell line HT-29 was purchased from the American Type Culture Collection. HT-29 cells were routinely grown in McCoy's 5A modified medium (Sigma) supplemented with antibiotics (penicillin (10 U/ml) and streptomycin (10 μg/ml)). All the cells were routinely maintained in 75 cm2 tissue culture flasks and incubated at 37°C in a humidified atmosphere (5 % CO2). The cells were passaged when the confluency of the flasks was approximately 80 %. For transcription analysis, the cells were seeded into 75 cm2 flasks and grown for 21 d with the medium being changed every other day. Before treatment, the cells were grown in antibiotic-free McCoy's 5A modified medium supplemented with 2 % fetal bovine serum for at least 24 h. During treatment, the cells were exposed to McCoy's 5A medium supplemented with BCO, HMO or 3′-sialyllactose (each 4 mg/ml) for 24 h. Non-supplemented McCoy's 5A medium was used as a control. After treatment, the cells were harvested from the flasks, washed twice with PBS and stored at − 80°C. Importantly, the oligosaccharides used in the present study did not affect the viability of the human cells as confirmed by simple trypan blue staining and real-time analysis of cell viability using an xCELLigence system (Roche) (data not shown).
Gene expression profiling
Human gene expression analysis
Total RNA from treated and untreated HT-29 cells was extracted using an RNA Extraction Mini kit according to the manufacturer's protocol. The quantity and purity of the RNA of each sample were determined by spectroscopy (A 260/A 280) and the size distribution was assessed using an Agilent Bioanalyzer (Agilent Technologies). Only samples with an A 260:A 280 ratio >1·8 were used during the analysis. Using the Low Input Quick Amp Kit (Agilent Technologies), 200 ng of total RNA were converted into labelled complementary RNA with nucleotides coupled to fluorescent dye Cy3 following the manufacturer's protocol. The purity and quantity of each sample were again determined by spectroscopy (A 260/A 280) and the size distribution was assessed using an Agilent Bioanalyzer. Cy3-labelled complementary RNA (600 ng) from each sample was hybridised to an Agilent Human GE 8x60k Microarray. The hybridised array was then washed and scanned, and data were then extracted from the scanned image using Feature Extraction version 10.7 (Agilent Technologies). All samples (no oligosaccharides, BCO, HMO and 3′-sialyllactose) were run on three separate occasions in triplicate.
Quantitative PCR arrays
To validate the expression of specific gene targets, quantitative PCR analysis was performed. Briefly, total RNA was isolated from treated and untreated HT-29 cells (1 × 106 cells) using a high pure RNA isolation kit (Roche) as per the manufacturer's instructions. RNA yield was determined by spectroscopy (A 260/A 280) and the integrity and size distribution of the RNA were determined on a denaturing agarose gel. The purity of the RNA was confirmed by PCR. The extracted RNA was reverse-transcribed using a first-strand cDNA synthesis kit (Roche), including 1 μg of total RNA and an oligo(dt) primer, as per the manufacturer's instructions. The reverse-transcribed product was amplified (RT-PCR) using a LightCycler® 480 instrument 96 (Roche) and the RealTime ready custom panels (Roche) as per the manufacturer's instructions. Briefly, each PCR contained 10 μl of LightCycler® 480 Probes Master, 9 μl of PCR-grade water and 1 μl of complementary DNA. The PCR running conditions were as follows: 10 min of initial denaturation step at 95°C followed by forty-five cycles of 10 s at 95°C, 30 s at 60°C and 1 s at 72°C, and a final cooling step of 40°C for 30 s. All samples (no oligosaccharides, BCO, HMO and 3′-sialyllactose) were run on three separate occasions in triplicate.
Data analysis
Gene expression analysis
To identify potential outliers and determine the overall similarity between replicate samples, individual hybridisations were subjected to a two-way agglomerative cluster analysis and a principal component analysis. The two-way agglomerative cluster analysis was carried out using Ward's minimum variance as the heuristic criterion and Pearson's correlation as the distance metric, together with average linkage as the heuristic criterion and cosine correlation as the distance metric. Intensity data for each replicate were generated into a single experiment using the Rosetta Resolver Agilent Intensity Experiment Builder pipeline with differentially expressed genes (P value < 0·05 and absolute fold change >1·5) detected using the Agilent-Ratio Builder. To determine the overall similarity between experiments (treatments), a principal component analysis and a two-way agglomerative cluster analysis were carried out.
To identify the biological processes regulated by each treatment, gene set enrichment analysis was performed using a Biological Networks Gene Ontology tool(Reference Maere, Heymans and Kuiper24). This tool constructed network diagrams illustrating the biological processes represented by the differentially expressed transcripts (DET). This construction involved the use of a hypergeometric statistical test with Benjamini and Hochberg false discovery rate multiple testing corrections to identify Gene Ontology terms that are overrepresented in DET found in the segregated groups. All the experiments were performed three times in triplicate, and representative data are presented. Where applicable, data are presented as means and standard deviations of replicate experiments.
RT-PCR
Relative quantification analysis was performed on replicate samples by comparing the expression level of the target gene of interest with the expression of a reference gene in a single sample. The final results are shown as the relative expression level of the normalised samples (Δ cycle threshold (C t)) in relation to the expression of the calibrator sample (2− ΔΔC t). The reference genes selected for this analysis included β-2-microglobulin, ribosomal phosphoprotein P0 and 18S rRNA. These genes were selected as they were shown to improve quantitative mRNA studies involving human intestinal epithelium(Reference Dydensborg, Herring and Auclair25). All the experiments were performed three times in triplicate, and representative data are presented. Where applicable, data are presented as means and standard deviations of replicate experiments, and Student's t test (P≤ 0·05) was used to determine whether the means were significantly different. Graphs were drawn using Microsoft Excel.
Results and discussion
Overall transcriptional response
In the present study, the transcriptional response of HT-29 cells to both HMO and BCO treatments was assessed using total RNA and Agilent Human GE microarrays. Initially, principal component analysis and unsupervised hierarchical clustering of the sample data were performed to demonstrate the good reproducibility between biological replicates (data not shown). DET were then detected by comparing the intensity data for the baseline treatment (no oligosaccharides) with those for the test treatment (oligosaccharides). The numbers of DET (P value < 0·05 and absolute fold change >1·5) for each treatment are presented as a Venn diagram in Fig. 1. The total number of DET for all the treatments was 2305. The HMO and BCO treatments shared 441 DET, and only 191 DET were shared by all the three treatments. The number of DET that were unique to the BCO treatment (n 602) was substantially higher than that of DET unique to the other treatments, indicating that the BCO treatment elicits a more widespread epithelial response than the other treatments. The high number of shared DET between the HMO- and BCO-treated samples suggests that the transcriptional responses of HT-29 cells to HMO and BCO share some degree of similarity. In support of this, the two-way agglomerative cluster analysis of the transcriptional data revealed that there were two major branches of sequence apparent, with the HMO and BCO treatments falling into one branch and the 3′-sialyllactose treatment falling into the other branch. It is notable that the number of shared transcripts comprised 37 % of the total number of HMO-induced transcripts but only 29 % of the BCO-induced transcripts. This would suggest that while there is a substantial overlap between the transcriptional responses induced by BCO and HMO, there is also a significant level of treatment-specific response.

Fig. 1 Venn diagram showing the number of differentially expressed transcripts shared by human milk oligosaccharide treatment (HMO), bovine colostrum oligosaccharide treatment (BCO) and 3′-sialyllactose (3′SL) treatment of HT-29 cells.
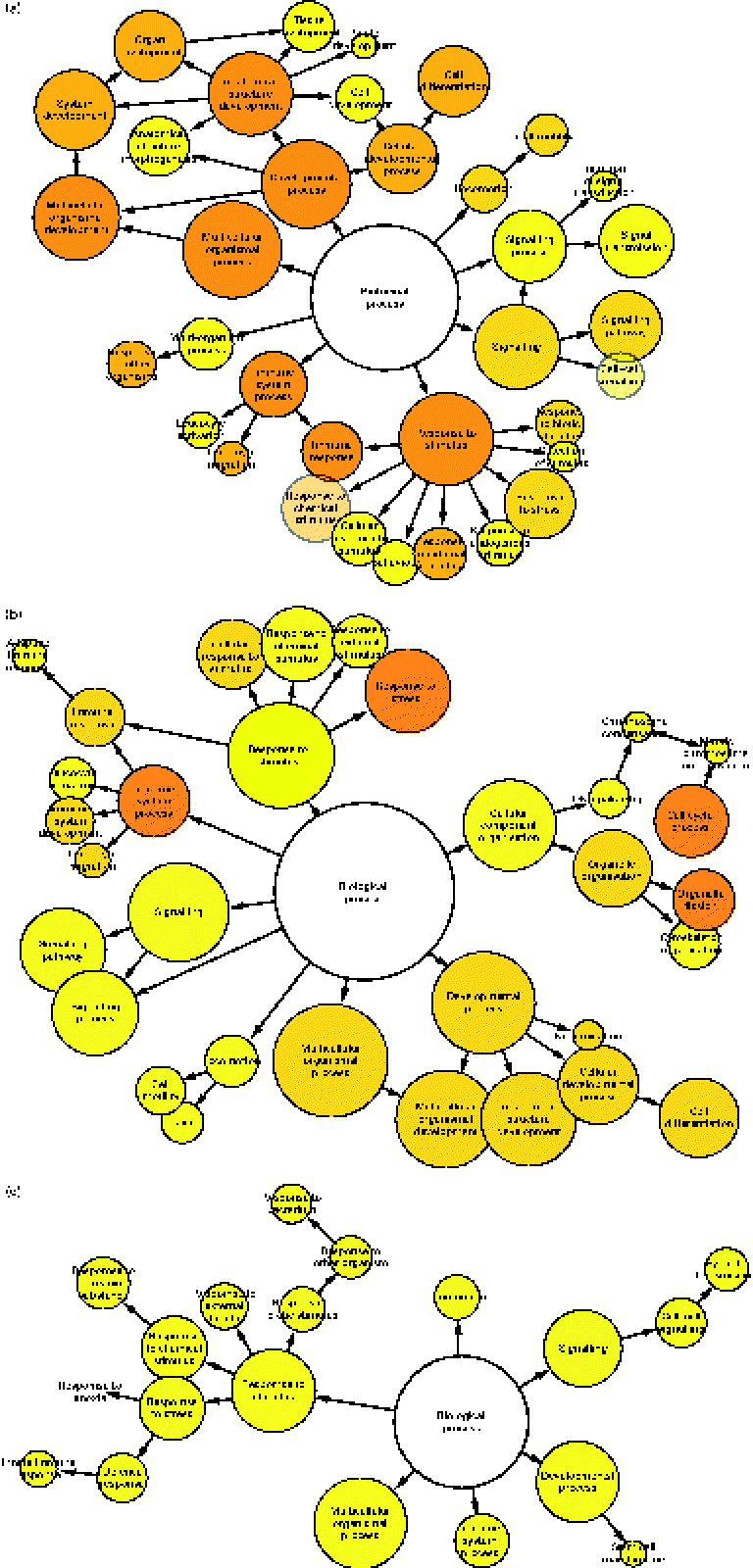
To determine which gene ontology terms were significantly overrepresented by the genes affected by each treatment, a gene set enrichment analysis was performed using a Biological Networks Gene Ontology tool. As illustrated in Fig. 2(a)–(c), network diagrams were constructed to reveal the biological processes that may be influenced by each treatment. All treatments had potential effects on numerous processes including a response to stimulus, signalling, locomotion, multicellular organismal processes, developmental processes and immune system processes. However, the significance (P value) of these gene ontology categories differed between each treatment (Table 1). Previously, HMO have been implicated in the regulation of some of these processes. For instance, they have been shown to directly influence developmental processes such as cell proliferation and differentiation. Kuntz et al. (Reference Kuntz, Rudloff and Kunz2) demonstrated that HMO inhibited the growth of HT-29, Caco-2 and HIEC cells in a concentration-dependent manner. Differentiation of HT-29 and HIEC cells was also enhanced. The ability of food-sourced glycans to induce phenotypic changes in host cells is probably due to the activation of cell signalling pathways, which can occur as glycan structures interact with receptors present on the surfaces of epithelial cells. This theory is supported by the high number of signalling genes differentially expressed after the HMO (149 genes), BCO (201 genes) and 3′-sialyllactose (ninety genes) treatments of HT-29 cells. Considering this, there is a need to identify such receptors in order to fully understand the functionality of oligosaccharides in the gut. To this end, Kuntz et al. (Reference Kuntz, Kunz and Rudloff26) demonstrated the ability of HMO to interact with the epidermal growth factor receptor on HT-29 cells and induce cell signalling, leading to cellular differentiation, proliferation and apoptosis. Interestingly, our data seem to support the findings of Kuntz et al. (Reference Kuntz, Rudloff and Kunz2), given that all treatments had a regulatory effect on developmental processes.

Fig. 2 Gene set enrichment analysis illustrating the biological processes regulated in HT-29 cells after treatment with (a) human milk oligosaccharide (HMO), (b) bovine colostrum oligosaccharide (BCO) and (c) 3′-sialyllactose (3′SL). Each node represents the number of genes that map to that gene ontology category, while the edges represent connections between the nodes. The size of each node represents the number of genes that map to the category represented by each node, and the colour saturation of the node represents the significance (P value) of the category represented by each node.
Table 1 Gene set enrichment analysis of the transcriptional response of HT-29 cells to milk oligosaccharides

HMO, human milk oligosaccharides; BCO, bovine colostrum oligosaccharides; 3′SL, 3′-sialyllactose.
The methodology used for the structural characterisation of the pooled BCO used in the present study has been published elsewhere(Reference Marino, Lane and Abrahams16). During this analysis, structural assignments for thirty-seven free glycans, including twenty sialylated species, were obtained by hydrophilic interaction liquid chromatography–HPLC coupled with exoglycosidase digestion and offline negative-ion-mode mass spectroscopy. A total of sixteen neutral structures were identified ranging from disaccharides to hexasaccharides including lactose, N-acetylgalactosaminyl glucose, N-acetyllactosamine, N, N′-diacetyllactosediamine, α3-N-acetylgalactosaminyl lactose, GlcNAc(β1-3)Gal(β1-4)Glc and Gal(β1-4)Gal(β1-4)Glc. Isoglobotriose was also identified as well as the isomers Gal(β1-6)Gal(β1-4)Glc and Gal(β1-3)Gal(β1-4)Glc. Other neutral oligosaccharides found to be present were 2′-fucosyllactose, GalNAc(α1-3)(Fuc(α1-2))Gal(β1-4)Glc, lacto-N-neotetraose, Gal(β1-4)GlcNAc(β1-6)(GlcNAc(β1-3))Gal(β1-4)Glc, lacto-N-novopentaose and lacto-N-neohexaose. Bovine colostrum was mainly composed of acidic oligosaccharides with the predominant structures being 3′-sialyllactose, 6′-sialyllactoseamine, 6′-sialyllactose and disialyllactose. High-molecular weight acidic structures included NeuNAc(α2-3)Gal(β1-4)GlcNAc(β1-6)(GlcNAc(β1-3))Gal(β1-4)Glc, NeuNAc(α2-6)Gal(β1-4)GlcNAc(β1-6) (GlcNAc(β1-3))Gal(β1-4)Glc and two sialylated forms of lacto-N-neotetraose, lacto-N-novopentaose and lacto-N- neohexaose. Other minor acidic oligosaccharides that were identified included the (α2-6) sialyl derivative of LacdiNAc, NeuNAc(α2-6)(GlcNAc(β1-3))Gal(β1-4)Glc, disialyllactosamine and two isomeric tetrasaccharides, NeuNAc(α2-3)Gal(β1-4)Gal(β1-4)Glc and NeuNAc(α2-6)(Gal(β1-3))Gal(β1-4)Glc. Finally, three structures containing the sialic acid variant, Neu5Gc, were identified (sialyllactose, sialyllactosamine and disialyllactose).
Structural characterisation of the HMO pool used in the present study is currently ongoing. Early findings correlate well with previous publications, where 100–150 oligosaccharides can be divided into two groups: (1) neutral oligosaccharides containing galactose, N-acetylglucosamine, fucose and lactose core and (2) acidic oligosaccharides containing the same oligosaccharide composition with additional N-acetylneuraminic acid(Reference Kobata and Ginsburg22, Reference Ninonuevo, Park and Yin27–Reference Boehm and Stahl31). Initial comparative analysis of the BMO and HMO pools revealed that they are composed of 90 % acidic oligosaccharides and 70 % neutral oligosaccharides, respectively. BCO that have not been reported to be present in human milk were identified as trisaccharides, containing (GalNAc(α1-3)Gal) units at the reducing end, as well as those containing Neu5Gc(Reference Marino, Lane and Abrahams16). Although there were distinct differences in the number and complexity of the oligosaccharides present in both pools, they shared a number of common structures such as lacto-N-neotetraose, 2′-fucosyllactose, 3′-sialyllactose and 6′-sialyllactose. These structures have been associated with many biological activities such as anti-infective activity, immunomodulation and cell differentiation(Reference Kuntz, Rudloff and Kunz2, Reference Mysore, Wigginton and Simon17, Reference Hakkarainen, Toivanen and Leinonen18), and their presence in both milks may explain the similarities observed between the HMO and BCO transcriptional responses in HT-29 cells. Indeed, these results may suggest that bovine colostrum is a sustainable source of biologically active oligosaccharides.
Overall, these results highlight the limited effect that a single glycan structure, in this case 3′-sialyllactose, can have on the transcriptome of human intestinal epithelia. For example, the number of DET affected after the HMO and BCO treatments of HT-29 cells was much higher than that affected after the 3′-sialyllactose treatment. Moreover, the significance of the gene ontology categories influenced by the 3′-sialyllactose treatment was lower than that influenced by the other treatments. Therefore, the food industry may need to consider the use of multiple oligosaccharide structures in functional foods in order to provide any health benefits associated with such ingredients. The use of mixtures of oligosaccharides may reduce costs in production as the fractionation of food-sourced oligosaccharides can often be both time consuming and expensive.
Regulation of immune system-associated glycogenes
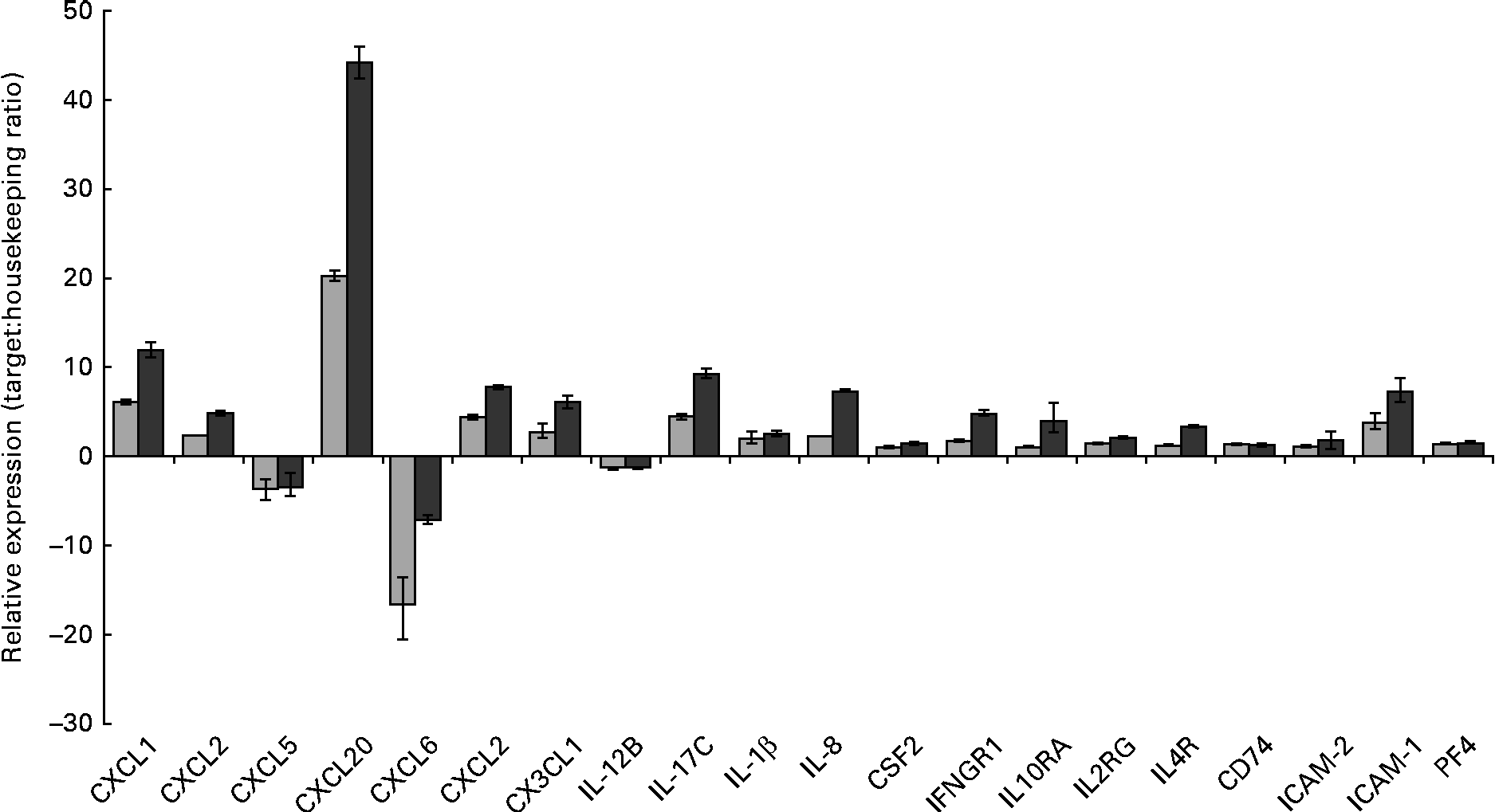
Gene set enrichment analysis of the transcriptional data revealed that the most significant (P value) biological process influenced by the HMO, BCO and 3′-sialyllactose treatments was an immune system response. To identify the immune system-associated glycogenes differentially expressed in the present study, the list of DET was compared with the human glycogene list (version 4) published by the Consortium for Functional Glycomics (Table 2). All the genes that were identified could be placed into three separate classes: cytokines, chemokines and cell surface receptors. To quantify and validate differences in the expression levels of these genes, we performed real-time PCR using complementary DNA and RealTime ready custom panels. As illustrated in Fig. 3, treatment of the HT-29 cells resulted in significant differences in the expression of the chemokines chemokine (C–X–C motif) ligand 1 (CXCL1), chemokine (C–X–C motif) ligand 3 (CXCL3), chemokine (C–C motif) ligand 20 (CCL20), chemokine (C–X–C motif) ligand 2 (CXCL2), chemokine (C–X–C motif) ligand 6 (CXCL6), chemokine (C–C motif) ligand 5 (CCL5), chemokine (C–X3–C motif) ligand 1 (CX3CL1) and CXCL2, as well as the cytokines IL-1β, IL-8, colony-stimulating factor 2 (granulocyte– macrophage) (GM-CSF2), IL-17C and the plate factor 4. Differential expression was also confirmed for cell surface receptor genes that code for interferon γ receptor-1, intercellular adhesion molecule-1 (ICAM-1) and -2 (ICAM-2) and IL-10 receptor α.
Table 2 Differential expression of the immune system-associated glycogenes

HMO, human milk oligosaccharides; BCO, bovine colostrum oligosaccharides; 3′SL, 3′-sialyllactose.
* No significant signal detected.
† Negative value.

Fig. 3 RT-PCR analysis demonstrating the changes in mRNA expression levels of immune-associated glycogenes after the human milk oligosaccharide (HMO, ![]() ) and bovine colostrum oligosaccharides (BCO,
) and bovine colostrum oligosaccharides (BCO, ![]() ) treatments of HT-29 cells. Error bars represent the range of possible relative expression values defined by the standard error of the delta threshold cycle (C t). CXCL1, chemokine (C–X–C motif) ligand 1; CXCL2, chemokine (C–X–C motif) ligand 2; CXCL5, chemokine (C–C motif) ligand 5; CXCL20, chemokine (C–C motif) ligand 20; CXCL6, chemokine (C–X–C motif) ligand 6; CX3CL1, chemokine (C-X3-C motif) ligand 1; CSF2, colony-stimulating factor 2 (granulocyte–macrophage); IFNGR1, interferon gamma receptor-1; IL10RA, IL10 receptor, alpha; IL2RG, IL2 receptor, gamma; IL4R, IL4 receptor; CD74, molecule, major histocompatibility complex, class II invariant chain; ICAM-2, intercellular adhesion molecule-2; ICAM-1, intercellular adhesion molecule-1; PF4, platelet factor 4.
) treatments of HT-29 cells. Error bars represent the range of possible relative expression values defined by the standard error of the delta threshold cycle (C t). CXCL1, chemokine (C–X–C motif) ligand 1; CXCL2, chemokine (C–X–C motif) ligand 2; CXCL5, chemokine (C–C motif) ligand 5; CXCL20, chemokine (C–C motif) ligand 20; CXCL6, chemokine (C–X–C motif) ligand 6; CX3CL1, chemokine (C-X3-C motif) ligand 1; CSF2, colony-stimulating factor 2 (granulocyte–macrophage); IFNGR1, interferon gamma receptor-1; IL10RA, IL10 receptor, alpha; IL2RG, IL2 receptor, gamma; IL4R, IL4 receptor; CD74, molecule, major histocompatibility complex, class II invariant chain; ICAM-2, intercellular adhesion molecule-2; ICAM-1, intercellular adhesion molecule-1; PF4, platelet factor 4.
Interestingly, exposure of the HT-29 cells to HMO and BCO resulted in the up-regulation of IL-17C and IL-1β. IL-17C belongs to the IL-17 family of cytokines whose members have been shown to induce the expression of pro-inflammatory chemokines and cytokines. Until recently, the biological function(s) of IL-17C were unknown. However, there is now evidence to suggest that IL-17C is an epithelial cell-derived cytokine with a function similar to that of IL-17A. Indeed, Pappu et al. (Reference Pappu, Ramirez-Carrozzi and Sambandam32) demonstrated the ability of IL-17C to act in an autocrine manner and induce a rapid innate immune response in epithelial cells. Moreover, it has been shown that IL-17C expression in colonic epithelial cells was induced by exposure to bacterial products (peptidoglycan and flagellin) or via the pro-inflammatory cytokine IL-1β. Thus, the up-regulation of IL-1β in the HT-29 cells after HMO and BCO treatments could account for the increased IL-17C expression. However, another contributing factor may be the presence of glycans that bind to the epithelial cell surface.
It is more than likely that the expression of IL-17C by the intestinal cells observed in the present study resulted in the up-regulation of other cytokine genes as well as chemokine genes. Previously, IL-17A has been shown to up-regulate the expression of a subset of chemokines and cytokines including CXCL1, chemokine (C–X–C motif) ligand 8 (CXCL8), CCL20, IL-6 and IL-8(Reference Lee, Wang and Kattah33). It has also been shown to negatively regulate the production of cytokines such as CCL5(Reference Lee, Wang and Kattah33). Similarly, Pappu et al. (Reference Pappu, Ramirez-Carrozzi and Sambandam32) demonstrated the ability of IL-17C to increase the expression of CXCL1, CXCL3, CCL20 and IL-8 as well as colony-stimulating factors in epidermal keratinocytes. We have observed that treating the HT-29 cells with BCO and HMO increased the expression of CXCL1, CXCL3, CCL20, CSF2 and IL-8 and decreased the expression of CCL5. Therefore, we hypothesise that in the early stages of infant development where both HMO and bacteria are highly abundant, these may induce the expression of IL-1β and IL-17C, leading to the production of pro-inflammatory cytokines and chemokines. Induction of the genes of these factors in the neonate epithelium in vivo would result in an epithelial innate immune response early in the postnatal period that could potentially protect against bacterial colonisation in infants. Interestingly, Pappu et al. (Reference Pappu, Ramirez-Carrozzi and Sambandam32) also demonstrated the unique ability of IL-17C to control homeostasis in the gut mucosa. Using a dextran sodium sulphate-induced colitis model, the researchers have demonstrated that the expression of IL-17C resulted in the resolution of the disease. IL-17C not only could provide protection during the early stages of life but may also play a significant role in the development and maturation of the infant intestinal immune response together with the establishment and maintenance of resident microflora. Other chemokines that were up-regulated in the presence of HMO and BCO were CXCL6 and CXCL2 (Table 2). In addition to their function as mediators of the innate immune response, several of the induced chemokines possess inherent antimicrobial activity. The chemokine gene with the highest level of induced transcription was CCL20, followed by CXCL1, CXCL6 and CXCL2, and these chemokines are known to have direct antimicrobial effects in addition to their role in immunomodulation(Reference Yang, Chen and Hoover34). Indeed, the human granulocyte chemotactic protein 2, CXCL6, is often expressed in epithelial cells during inflammation and displays antibacterial activity against a number of enteric bacteria, possibly protecting against bacterial colonisation of the intestinal tract.
Interestingly, there is emerging evidence that CCL20 is consistently induced by epithelial cells in response to probiotic bacteria such as Lactobacillus salivarius (Reference Ryan, O'Hara and van Pijkeren35–Reference O'Callaghan, Butto and Macsharry37) and Lactobacillus acidophilus (Reference O'Flaherty and Klaenhammer38). The expression of pro-inflammatory genes can lead to the up-regulation of the NF-κB complex, resulting in intestinal inflammation, which can be an undesirable response in the host if it results in a chronic state. The pro-inflammatory potential of such genes would be attenuated by the induction of a number of genes known to have an anti-inflammatory effect on the NF-κB transcription factor. The most significant of these are baculoviral IAP repeat containing 3 (BIRC3), NF-κ light polypeptide gene enhancer in B-cells inhibitor, α (NFKBIA), NF-κ light polypeptide gene enhancer in B-cells inhibitor, ɛ (NFKBIE) and TNFα-induced protein 3 (TNFAIP3). These negative regulators act by sequestering NF-κB in the cytoplasm, preventing it from reaching the nucleus, and ensuring that the NF-κB complex is poised in favour of an anti-inflammatory response even in the presence of pro-inflammatory chemokines. As with the increased transcription of chemokines, similar up-regulation of the NF-κB regulatory genes has been reported for a number of probiotic bacteria, both in vitro (Reference O'Callaghan, Butto and Macsharry37, Reference O'Flaherty and Klaenhammer38) and in vivo (Reference van Baarlen, Troost and van der Meer39, Reference van Baarlen, Troost and van Hemert40). Therefore, there is accumulating evidence to suggest that many probiotic bacteria can cause the NF-κB complex to shift in favour of an anti-inflammatory-type response in which the complex is sequestered in the cytoplasm but retains the potential to trigger an inflammatory response when required. The potential to activate measured and proportionate pro- and anti-inflammatory responses is crucial as complete inactivation of NF-κB can result in a pleiotropic dysfunction of cellular processes(Reference Spehlmann and Eckmann41). What is less well understood is the effect of milk oligosaccharides on epithelial immune responses. There have been some reports that human milk constituents may down-regulate the inflammatory response in intestinal epithelial cells(Reference Claud, Savidge and Walker42, Reference Daddaoua, Puerta and Requena43); however, this hypothesis requires further validation. Interestingly, our data demonstrated the up-regulation of both pro-inflammatory genes and NF-κB regulatory genes in the presence of milk oligosaccharides, which while causing a generalised immune system response, is likely to direct the NF-κB complex towards an anti-inflammatory response. The up-regulation of the regulatory RNase Zn finger CCCH-type containing 12A (ZC3H12A) is interesting as this has a significant role in the modulation of innate immune responses, particularly of those associated with autoimmune dysfunction(Reference Matsushita, Takeuchi and Standley44). The data presented here raise the possibility that in addition to their well-recognised role in the physical blocking of pathogenic bacteria, milk oligosaccharides may also act directly on the host immune system to induce a host-derived antimicrobial response and also reduce intestinal inflammation.
As illustrated in Table 2, BCO treatment of the HT-29 cells altered the expression of a number of immune system-associated cell surface receptors, some of which were not expressed after the HMO treatment. Indeed, the up-regulation of the intercellular adhesion molecule ICAM-1 was the only mechanism shared by both treatments. These receptors could function early in the postnatal period by interacting with breast milk components. Human breast milk contains its own immune system components including cytokines, macrophages, neutrophils and lymphocytes. Indeed, colostral (approximately 4 × 109 per litre) and mature (approximately 1 × 108–109 per litre) milk leucocytes are composed of 55–60 % macrophages, 30–40 % neutrophils and 5–10 % lymphocytes(Reference Field45). These components in milk provide protection for suckling infants while also facilitating immune system development and maturation. For example, the maternal cytokines transforming growth factor β, IL-6 and IL-10 have been shown to contribute to the differentiation of IgA-producing cells and the development and maturation of the naive intestinal immune system(Reference Field45). The induction of cell surface receptors by HMO and BCO could be important to the health of suckling infants. For example, ICAM-1 functions during the transepithelial migration of leucocytes by interacting with lymphocyte function-associated antigen 1 (LFA-1) expressed on the surface of circulating leucocytes. Leucocytes provide protection for infants by neutralising any potential danger on-site. However, excessive leucocyte infiltration can lead to the emergence of an inflammatory disease. Therefore, this process must be controlled physiologically. Previously, Bode et al. (Reference Bode4) demonstrated the ability of acidic HMO to bind to circulating leucocytes and thereby reduce rolling and adhesion. 3′-Sialyllactose was identified as one of the major inhibitory components in this work. Considering this, HMO and BMO could result in a controlled leucocyte response in the gut by countering the expression of ICAM-1 with anti-adhesive activity.
Cell surface receptor genes that were only up-regulated in the presence BCO included interferon γ receptor-1 (IFNGR1), ICAM-2 and IL-10 receptor α (IL10RA). The glycoprotein IFNGR1 is predominantly responsible for mediating high-affinity, species-specific ligand binding, ligand trafficking and signal transduction. It has been shown to increase natural resistance to bacterial, parasitic and viral infections. Therefore, the expression of IFNGR1 could provide increased protection in newborns. ICAM-2 is mainly found on resting endothelial cells, but it has been shown to be expressed by epithelial cells. It contributes to lymphocyte recirculation by blocking LFA-1-dependent cell adhesion. Thus, ICAM-2 functions to control the infiltration of leucocytes. IL10RA is a cell surface receptor of the cytokine synthesis inhibitory factor IL-10, which can inhibit the synthesis of pro-inflammatory cytokines interferon γ (IFN-γ), IL-2, IL-3, TNF-α and GM-CSF as well as stimulate T-cell and B-cell maturation. The interaction of these receptors with their respective ligands, which have been shown to be present in human milk, could result in a controlled immune response in infants. Moreover, their presence could enhance immune competence while also contributing to the maturation of the intestinal immune response.
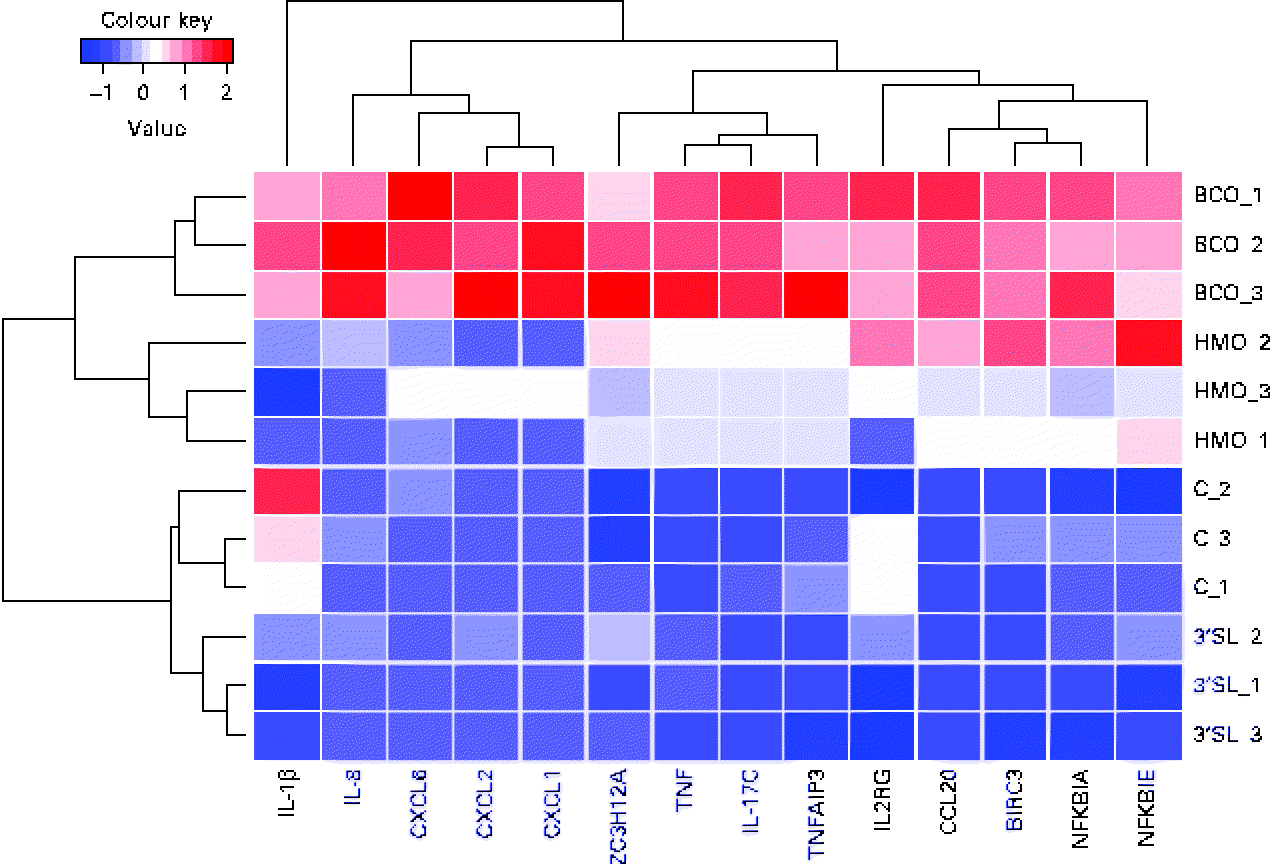
In the global analysis of DET levels described above, we discussed the overlap observed between the different treatments and the substantial difference between the number of DET induced by BCO and HMO. To investigate the hypothesis that there may be a differential regulation of immune system gene transcription induced by the different treatments, we examined the transcriptional patterns of the immune system genes described in the present study. It is clear from Fig. 4 that, as was observed for the global transcriptional pattern resulting from the BCO and HMO treatments, the immune system gene transcription levels could be segregated into four clusters, each corresponding to a single set of experimental replicates. The BCO and HMO replicates formed a separate grouping when compared with the 3′-siallylactose and control samples. The overall transcript levels of the immune system genes for the HMO and BCO treatments were substantially higher than those for the 3′-siallylactose and control treatments. Further separation between the BCO and HMO samples was observed when considering the transcript levels of the cytokines CXCL1, CXCL2, CXCL6, IL-8 and IL-1β. As has been discussed previously, the BCO and HMO samples share a number of common oligosaccharide structures; however, these structures are present in each sample at different concentrations, which could account for differences in the transcript levels of these immune system-associated genes. It is also notable that the clustering of genes resulted in three distinct groups that could be designated as containing mainly cytokine and chemokine, NF-κB-related and immune system genes with multiple functions (ZC3H12A, TNF, IL-17C and TNFAIP3). Overall, the data shown in Fig. 4 are in agreement with the inferences drawn from Fig. 1, indicating that BCO and HMO are more effective in inducing transcriptional changes when compared with 3-siallylactose, with HMO having the most substantial effect. However, there was no evidence to suggest that a significant pro-inflammatory response was induced. In fact, the pattern of immune system genes induced by BCO was similar to that induced by HMO, with the only difference being that the transcript levels were generally higher following the BCO treatment. Thus, the effect of BCO on the immune system is likely to be at least as beneficial as that of the HMO.

Fig. 4 Heatmap and cluster analysis of transcriptional changes induced in a group of immune system-related genes in HT-29 epithelial cells following treatment with bovine colostrum oligosaccharides (BCO), human milk oligosaccharides (HMO) and 3′-sialyllactose (3′SL). Gene transcription in untreated cells (C) is included for reference. CXCL6, chemokine (C–X–C motif) ligand 6; CXCL2, chemokine (C–X–C motif) ligand 2; CXCL1, chemokine (C–X–C motif) ligand 1; ZC3H12A, Zn finger CCCH-type containing 12A; TNFAIP3, TNFα-induced protein 3; IL2RG, IL2 receptor, gamma; CCL20, chemokine (C–C motif) ligand 20; BIRC3, baculoviral IAP repeat containing 3; NFKBIA, NFκ light polypeptide gene enhancer in B-cells inhibitor, α; NFKBIK, NFκ light polypeptide gene enhancer in B-cells inhibitor, ɛ.
Concluding remarks
The lower incidence of infectious and inflammatory diseases in breast-fed infants than in formula-fed infants has resulted in efforts being made to decipher the beneficial ingredients in human milk. In the present study, we highlighted the potential of HMO as bioactive ingredients that could influence many biological processes in colonic epithelial cells. We also highlighted the promise of bovine colostrum as a suitable source of oligosaccharides that may function similarly to those found in human milk. We demonstrated the ability of HMO and BCO to increase the level of expression of cell surface receptors, chemokines and cytokines in HT-29 cells. Specifically, these oligosaccharides induced the expression of an epithelial cell-derived cytokine, IL-17C. The expression of IL-17C in vivo could result in increased protection for neonates while also contributing to the maturation of the intestinal immune response. Indeed, the consumption of HMO and BCO in the postnatal period may promote the maturation of the naive cytokine response and contribute to lower immune competence in infants. Although there were similarities observed between the HMO and BCO treatments, there were distinct differences such as the number and induction level (P value) of DET. These differences are more than likely due to the limited number and varying concentrations of common oligosaccharides in each sample. Considering these results, further studies are required to determine whether these differences have varying effects on human health. Furthermore, there is a need to investigate whether these transcriptional changes can result in differential phenotype expression. In conclusion, our data highlight the potential of food-sourced glycans to induce a pro-inflammatory response in colonic epithelial cells, which could play a significant role in the development and maturation of the intestinal immune response.
Acknowledgements
The present study was funded by the Department of Agriculture and Food, Ireland, under the Food Institutional Research Measure, project reference no. 05/R&D/TD/368, and Science Foundation Ireland Alimentary Glycoscience Research Cluster (AGRC), grant no. 08/SRC/B1393. The authors' contributions are as follows: S. D. C., R. M. H. and J. A. L. conceptualised and designed the study; J. A. L. conducted the experiments and acquired the data; J. A. L. and J. O'C. analysed and interpreted the data; J. A. L. wrote the first draft of the manuscript; S. D. C., R. M. H., J. O'C. and J. A. L. critically revised the manuscript and are primarily responsible for the final content. The authors declare that there are no conflicts of interest.