Coeliac disease (CD) is an immune-mediated enteropathy characterised by chronic inflammation of the proximal small intestine caused by ingestion of gluten in genetically predisposed individuals who carry the HLA-DQ2 or the DQ8 gene( Reference Ludvigsson, Leffler and Bai 1 ). Although 30 % of the general population carries the genetic predisposition for CD, only approxmately 3 % will develop this disease. It has been speculated that additional environmental factors may be required. Several studies have reported an association between abnormal intestinal microbiota composition (dysbiosis) and CD( Reference Nistal, Caminero and Vivas 2 – Reference Collado, Donat and Ribes-Koninckx 5 ), but the underlying mechanism has not been demonstrated yet. A strict life-long gluten-free diet (GFD) is the only efficient treatment available for CD to date( Reference Freeman 6 ).
Gluten is a common component in the Western diet. Nevertheless, gluten is the main environmental trigger of CD, one of the most common food intolerances in humans, affecting approximately 1:100 individuals in Western countries. In vitro studies have shown that gluten proteins, due to their high proline content (approximately 15 %), are resistant to complete digestion by human digestive enzymes in the small intestine as those enzymes are deficient in prolyl endopeptidasic activity. As a result, many high molecular weight oligopeptides (10 to ≥30 amino acids residues) persist in the lumen of the small intestine and some of them, such as 33-mer or 19-mer, are capable of triggering the inflammatory process associated with CD( Reference Shan, Molberg and Parrot 7 , Reference Shan, Qiao and Arentz-Hansen 8 ). It has been described that dietary components not digested by human enzymes serve as substrates for intestinal bacteria. In fact, there is evidence to suggest that the human microbiota is involved in metabolism of gluten proteins; however, this issue is often underestimated( Reference Davila, Blachier and Gotteland 9 , Reference Caminero, Nistal and Herrán 10 ). Recent studies have shown that the oral cavity and the colon are colonised by bacteria that produce proteases capable of hydrolysing gluten proteins. Some of these bacteria are able to hydrolyse the non-digested peptides that trigger CD( Reference Zamakhchari, Wei and Dewhirst 11 – Reference Caminero, Nistal and Arias 13 ). Recently, our group has described the culturable microbiota involved in the metabolism of gluten proteins in the human gut( Reference Caminero, Herrán and Nistal 14 ). Interestingly, these bacteria are some of those that have been reported to be altered in CD patients, such as Bifidobacterium, Lactobacillus, Bacteroides, Staphylococcus, Clostridium and Escherichia coli ( Reference Sanz, De Pama and Laparra 15 ).
Despite the ubiquity of wheat in the diet and the importance of gluten proteins in the pathological process of CD, there are few studies that have focused on the relationship between gluten metabolism and gut microbiota in humans. Accordingly, the objective of this study was to analyse the metabolic activity of intestinal bacteria associated with gluten intake in healthy individuals, CD patients and first-degree relatives of CD patients.
Methods
Subjects
A total of twenty-two untreated CD patients (mean age 39·5 years, range 15–60 years), twenty treated CD adults on a GFD for at least 1 year (mean age 30·6 years, range 22–42 years), sixteen healthy volunteers on normal diet (mean age 30·1 years, range 25–45 years), eleven healthy volunteers on GFD (mean age 32·2 years, range 25–45 years), seventy-one first-degree relatives of CD patients on normal diet (mean age 36·4 years, range 24–59 years) and sixty-nine relatives on GFD (mean age 36·2 years, range 24–59 years) were enrolled for this study (online Supplementary Table S1).
The inclusion and exclusion criteria of the volunteers were as follows: (i) healthy volunteers in whom CD and other organic diseases of the gastrointestinal tract were ruled out; (ii) CD patients who were diagnosed by positive tTGA antibodies, HLA-DQ2 or DQ8 phenotype and a positive duodenal biopsy (>Marsh IIIa), treated CD patients recovered the normal architecture of the duodenum and the normal tTGA levels after a GFD for at least 1 year; and (iii) first-degree relatives of CD patients in whom CD was ruled out (no damage in duodenal biopsy and normal tTGA levels); fifty-eight of these volunteers showed HLA-DQ2 or HLA-DQ8. Relatives were on a GFD for 1 month, but two volunteers were not included because they did not follow the correct diet. Recruitment of healthy volunteers on a reliable GFD was not easy and we had to reduce the GFD to 1 week in this group. Volunteers were instructed by our staff to follow a correct GFD. All the volunteers filled out dietary quizzes to detect possible transgressions on GFD. None of the volunteers included in this study had been treated with antibiotics for at least 2 months before the sampling time.
The number of faecal samples used in each assay was different due to different reasons: some faecal samples did not have enough quantity to perform all the studies, some samples showed problems in specific assays and expensive experiments such as 33-mer proteolysis and faecal gluten quantification were limited due to economic reasons.
This study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all the procedures involving human subjects were approved by the local ethics committee of our hospital. Written informed consent was obtained from all the subjects.
Faecal sampling
All the faecal samples were freshly processed within 3 h of collection. For enzymatic determinations, faecal samples were thoroughly mixed with saline (1:5), maintained at 4°C for 90 min and centrifuged at 8000 rpm; the pellets were discarded after centrifugation. Aliquots were stored at –80°C or –20°C for later analysis.
Faecal tryptic activity and faecal glutenasic activity
Faecal tryptic activity (FTA) is a specific proteolytic activity like the trypsin proteolytic activity measured in faeces and is used to study bacterial metabolism. Faecal glutenasic activity (FGA) is a non-specific proteolytic activity towards gluten proteins excreted in the faeces( Reference Caminero, Nistal and Arias 13 ). Assays for FTA and FGA were performed following the protocol described by Caminero et al.( Reference Caminero, Nistal and Arias 13 ).
Faecal prolyl endopeptidasic activity
Prolyl endopeptidases are enzymes that cleave peptides with internal proline residue from the carboxyl side( Reference Shan, Molberg and Parrot 7 ). Faecal prolyl endopeptidasic activity (FPEP) activity was measured using fluorimetric method( Reference Goossens, De Meester and Vanhoof 16 ). A 10 µl supernatant fraction was diluted in 100 µl of K-phosphate buffer (100 mm) (pH 7·5), 1 mm-EDTA and 1 mm-DTT, pre-incubated at 37°C. The reaction was initiated by adding of 5 µl substrate (0·12 mm). The substrate used was N-benzoyloxycarbonyl-glycyl-prolyl-7-amino-4-methylcoumarin (dissolved in DMSO). The incubation time was 120 min at 37°C. The reaction was terminated with acetic acid, and the fluorescence was measured at 370 nm excitation and 440 nm emission wavelength. Enzymatic activities were calculated from a standard curve established using 7-amino-4-methylcoumarin. One unit of enzyme activity was defined as the amount of enzyme catalysing the formation of 1 µmol of 7-amino-4-methylcoumarin/min (IU) per kg of faeces.
Faecal dipeptidyl peptidasic IV activity
Faecal dipeptidyl peptidasic IV activity (FDPPIV) is an enzymatic activity that hydrolyses peptides with internal prolyl bonds from the N-terminal. FDPPIV activity was analysed by colorimetric determination of glycyl-prolyl-p-nitroanilide tosylate hydrolysis( Reference Nagatsu, Hino and Fuyamada 17 , Reference Elgün, Keskinege and Kumbasar 18 ). A 10 µl supernatant fraction was added to 100 µl of glycine–NaOH buffer (71 mm) (pH 8·5). The reaction was initiated by adding 5 µl of the substrate glycyl-l-proline p-nitroanilide p-tosylate (3 mm). The reaction took place at 37°C and was terminated after 30 min by adding 50 µl of 1 m-acetic acid. The absorbance of the p-nitroanilide liberated was measured at 385 nm. Enzymatic activities were calculated from a standard curve established using 4-nitroaniline. One unit of enzyme activity was defined as the amount of enzyme catalysing the formation of 1 µmol of p-nitroaniline/min (IU) per kg of faeces.
Determination of 33-mer hydrolysis
A synthetic 33-mer peptide (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) was synthesised with a purity of 95 % (Proteogenix). The reaction mixtures, (60 µl) containing 5 µl of faecal supernatant fraction and 60 µm of 33-mer peptide in PBS (pH 7·3), were incubated at 37°C for 24 h. The reactions were terminated by incubation at 100°C for 10 min, and 40 µl of each reaction sample was subjected to reverse-phase HPLC using a C-18 column (Lichrospher 100 RP18 column 5 µm, 4×250 mm; Teknokroma SL). The elution phases consisted of (a) MilliQ H2O containing 0·1 % trifluoroacetic acid (v/v) and (b) acetonitrile and 0·1 % trifluoroacetic acid (v/v). The eluate was monitored by UV absorbance at 215 nm( Reference Caminero, Herrán and Nistal 14 ).
Faecal SCFA
SCFA are metabolites produced when components from the diet are fermented by bacteria in the large intestine. SCFA were analysed as described by Caminero et al.( Reference Caminero, Nistal and Arias 13 ). In brief, 1 g of faeces was suspended in 2 ml of water and homogenised in a vortex mixer for 2 min. The suspension was centrifuged at 12 000 rpm, and the pH of the supernatant was decreased by adding HCl. The internal standard 4-methylvaleric acid solution was spiked into the supernatant (final concentration 258 µm). Following this, the supernatant was injected into a GC for analysis. Chromatography was performed in the LTI laboratory at the Universidad de León. The SCFA were identified on chromatograms according to the procedure described by Zhao et al.( Reference Zhao, Nyman and Jönsson 19 ). To quantify the peak area in terms of concentration, the relative response factor was used, following the methodology described by Ranilla et al.( Reference Ranilla, Carro and López 20 ).
Study of cultivable bacteria involved in gluten metabolism
Cultivable bacteria involved in gluten metabolism were characterised as described by Caminero et al.( Reference Caminero, Herrán and Nistal 14 ). In brief, faecal samples were cultured in two ways: (1) plated directly on solid MCG-3 after being diluted in saline and (2) inoculated in liquid MCG-3 medium, and after incubation at 37°C an aliquot of serial dilutions from each liquid culture was plated on MCG-3. Bacteria were isolated based on colony morphology. All the colonies were tested for glutenase activity (extracellular proteolytic activity determined by the presence of a halo surrounding the colony) and the ability to metabolise gluten (no growth on the isolation medium without gluten). Only the bacteria positive for any of these tests were identified by partial sequencing of the 16S rDNA gene using the universal primers 27F and E939R( Reference Nistal, Caminero and Vivas 2 ).
Denaturing gradient gel electrophoresis
Bacterial communities grown in liquid MCG-3 were studied by DGGE. Genomic DNA from liquid media was obtained using SpeedTools Tissue DNA Extraction kit (Biotools), and PCR fragments of 200 bp were amplified using the universal primers HDA1-GC and HDA2( Reference Nistal, Caminero and Vivas 2 ). DGGE analysis of the PCR amplicons was performed using the DCode Universal Mutation Detection System (Bio-Rad). The linear denaturing gradient of urea and formamide used for the separation of the amplicons was 35–55 %. Selected DGGE bands were re-amplified with the corresponding primers without the GC clamp, and the resulting PCR products were cloned with the StrataClone PCR cloning kit (Stratagene) and sequenced. The similarities between the banding patterns generated by PCR-DGGE analysis were analysed using the Dice coefficient and the unweighted-pair group method with the arithmetic average (UPGMA) clustering algorithm and were shown graphically as a dendrogram( Reference Nistal, Caminero and Vivas 2 ).
Gluten quantification
The amount of gluten in faeces and in liquid MCG-3 media after incubation with faeces was measured using the Competitive ELISA GlutenTox Kit (Biomedal) according to the manufacturer’s protocol. This Competitive ELISA contains G12 antibody that recognises a frequent epitope within the 33-mer peptide. G12 searches for the QPQLPY sequence found only in the 33-mer peptide from the α-gliadin protein. This kit is especially indicated for gluten quantification in faecal samples.
Statistics
All the variables were analysed using SPSS v. 18.0. Categorical variables were expressed as numbers and percentages, and quantitative variables were expressed as means and standard deviations. The ANOVA test was performed to evaluate differences between various samples. The Student’s t test and Wilcoxon’s test were performed to evaluate the differences between two independent and related samples, respectively. A P-value <0·05 was selected to reject the null hypothesis by two-tailed tests and a Bonferroni correction was applied. Pearson’s correlation coefficient was used to analyse whether the different parameters were related.
Results
Faecal glutenasic activity and faecal tryptic activity are increased in adult coeliac disease patients
A total of 209 human faecal samples belonging to twenty-two untreated CD patients, twenty treated CD patients, sixteen healthy volunteers on normal diet, eleven healthy volunteers on GFD, seventy-one relatives on normal diet and sixty-nine relatives on GFD were tested for FTA, FGA, FPEP and FDPPIV. The results obtained are represented in Fig. 1. The differences were statistically significant for FTA between CD patients v. healthy volunteers and CD patients v. first-degree relatives on normal diet and on GFD. In contrast, FGA was higher in CD patients than in healthy volunteers and relatives of CD patients on GFD (Fig. 1). Significant differences were observed when we compared healthy volunteers and relatives on GFD with treated CD patients. No differences were found between groups in FDPPIV and FPEP activities or when samples were analysed according to HLA-DQ2/8 status (data not shown).

Fig. 1 Values of faecal glutenasic activity (FGA), faecal tryptic activity (FTA), faecal dipeptidyl peptidase IV (FDPPIV) and faecal prolyl endopeptidase (FPEP) in healthy volunteers on normal diet (HND) and gluten-free diet (HGFD), untreated coeliac disease (CD) patients (UCD), treated CD patients (TCD) and first-degree relatives on normal diet (RND) and gluten-free diet (RGFD). Continuous bars show significant differences related to CD diagnostics. Dashed bars show significant differences related to gluten-free diet. Black circles indicate outliers.
The concentration of faecal SCFA is increased in adult coeliac disease patients and first-degree relatives
In all the faecal samples, acetic acid was the main SCFA, followed by propionic and butyric acids. The results showed that in CD patients and first-degree relatives, the concentration of faecal SCFA was higher compared with healthy volunteers. The differences were statistically significant for the concentrations of acetic acid, propionic acid, butyric acid and total SCFA between the healthy group v. the untreated CD patients group and between the healthy group and the relatives group when all of them were on normal diet. Relatives on a normal diet also showed significant differences in valeric acid contents when compared with healthy volunteers on normal diet. Significant differences in acetic acid, propionic acid, n-butyric acid, n-valeric acid and total of SCFA were observed between healthy v. treated CD patients and healthy v. relatives when all of them were on a GFD. Moreover, first-degree relatives on GFD had higher concentrations of isovaleric acid compared with healthy volunteers on GFD and a higher concentration of branched SCFA (isobutyric acid and isovaleric acid) than treated CD patients (Table 1).
Table 1 SCFA content in faecal samples of sixteen healthy volunteers on normal diet (ND), eleven healthy volunteers on gluten-free diet (GFD), twenty-two untreated coeliac disease patients, eighteen treated coeliac disease patients, seventy-one first-degree relatives of coeliac disease patients on normal diet and fifty-seven relatives on GFD (Mean values and standard deviations)

* Mean of samples (mmol/kg faeces).
† Significant differences v. healthy volunteers in ND (P<0·05).
‡ Significant differences v. healthy on GFD (P<0·05).
§ Significant differences v. coeliac disease patients on GFD (P<0·05).
Faecal samples showed peptidasic activity towards the 33-mer peptide
33-mer is an immunodominant peptide generated as a result of the incomplete digestion of gluten proteins carried out by the human digestive proteases. The presence of peptidasic activity against the 33-mer peptide was tested in seventy randomised human faeces samples from fifteen healthy volunteers on normal diet, eleven healthy volunteers on GFD, thirteen untreated CD patients, twenty treated CD patients, and eleven relatives on normal diet; 57 % of the faecal supernatants had the ability to completely hydrolyse this peptide. The chromatogram generated after the hydrolysis of 33-mer was characteristic of each sample (Fig. 2), suggesting a microbial origin of this peptidasic activity. We also performed a comparative study of the faecal peptidasic activity based on genetics, CD diagnostics and diets. None of the volunteers showed a variation in the peptidasic activity against 33-mer peptide after GFD (Fig. 3(a)). Among the faecal samples with peptidasic activity, those from CD patients hydrolysed the 33-mer peptide faster than samples from non-CD subjects (Fig. 3(a)), and this result was statistically significant.

Fig. 2 Examples of chromatograms generated after the incubation of the peptide 33-mer with different faecal samples for 2, 6, 8 and 12 h. The peptide 33-mer appears in minute 13 approximately AU, arbitrary units.

Fig. 3 (a) Degradation time of 33-mer peptide in healthy volunteers on normal diet (HND) and gluten-free diet (HGFD), untreated coeliac disease patients (UCD), treated coeliac disease patients (TCD) and first-degree relatives on normal diet (RND). (b) Correlation between faecal glutenasic activity (FGA) and degradation time of 33-mer peptide in faeces. ![]() , Healthy people on normal diet;
, Healthy people on normal diet; ![]() , Healthy people on gluten-free diet;
, Healthy people on gluten-free diet; ![]() , Untreated coeliac disease patients;
, Untreated coeliac disease patients; ![]() , Treated coeliac disease patients;
, Treated coeliac disease patients; ![]() , First-degree relatives on normal diet.
, First-degree relatives on normal diet.
Correlation between faecal glutenasic activity and the peptidasic activity towards the 33-mer
We studied whether proteolytic activities detected in faeces were related using Pearson’s correlation coefficient. No significant correlation was detected between FGA, FTA, FPEP and FDPPIV. However, a correlation between FGA and peptidasic activity towards 33-mer was found. Faecal samples with high FGA hydrolysed the 33-mer peptide faster than samples with low FGA that usually did not degrade this peptide (Fig. 3(b)).
The amount of gluten in faeces is influenced by gluten intake and the diagnosis of coeliac disease
The amount of gluten excreted in faeces was quantified in samples from fourteen healthy volunteers on normal diet, eleven healthy volunteers on GFD, eighteen untreated CD patients, fifteen treated CD patients and thirty-eight relatives on normal diet. The amount of gluten was higher in samples obtained from normal diet groups. The highest amount was found in faecal samples from healthy volunteers. Significant differences in normal diet were found between healthy volunteers v. untreated CD patients and relatives v. untreated CD patients, suggesting that the disease can affect gluten excretion. All the groups on GFD that were analysed had reduced amounts of gluten in faeces with values close to nil. Significant differences were also observed in healthy volunteers and CD patients between normal diet v. GFD that seem to be related to the diet. Furthermore, significant differences based on diagnostics appeared between treated CD patients v. healthy volunteers on GFD. (Fig. 4(a)).

Fig. 4 (a) Amount of gluten excreted by healthy volunteers on normal diet (HND), healthy volunteers on gluten-free diet (HGFD), untreated coeliac disease patients (UCD), treated coeliac disease patients (TCD) and first-degree relatives on normal diet (RND). Continuous bars show significant differences related to coeliac disease (CD) diagnostics. Dashed bars show significant differences in relation to gluten-free diet. (b) Relation between the amounts of gluten excreted in the faeces and faecal glutenasic activity (FGA) measured in samples from volunteers on ND. Black circles represent outliers. ppm, Parts per million.
Faecal glutenasic activity affects the concentration of gluten proteins in faeces
We studied whether proteolytic activities were related to gluten excretion in faeces. An inverse correlation between the amount of gluten in faeces and the different proteolytic activities measured in faeces has been found. Although no significant correlation appeared, the results showed that gluten excretion in faeces was influenced by FGA. Fig. 4(b) shows that the samples with high FGA, independent of the diagnosis, had a very small amount of gluten in faeces – that is, faeces with FGA higher than 10 000 U/g of faeces had scarce amounts of gluten. Significant differences were found between samples with a reduced activity (<4000 U/g faeces) v. faeces with an intermediate (4000–10 000 U/g faeces) or a high FGA (>10 000 U/g faeces) (Fig. 4(b)). In order to study this correlation in vivo, seven volunteers with low FGA and seven volunteers with high FGA (without CD diagnosis) were put on a high-gluten-containing diet for 1 month. Volunteers were on their normal diet supplemented with 15 g gluten/d for 1 month and after that faecal samples were collected. Gluten was consumed through bread slices. Results showed a significant lower amount of gluten in volunteers with a high FGA compared with volunteers with a low FGA (Fig. 5).

Fig. 5 Differences in faecal gluten between volunteers with a low faecal glutenasic activity (FGA) and volunteers with a high FGA on a high-gluten-containing diet. Volunteers were on their normal diet supplemented with 15 g gluten/d for 1 month. ppm, Parts per million.
Study of bacteria involved in gluten metabolism showed significant changes in Lactobacillus and Clostridium genera
In order to characterise bacteria involved in gluten metabolism, faecal samples from seven healthy volunteers, five untreated CD patients and ten first-degree relatives were cultured in MCG-3 medium. Bacteria were selected based on the ability to metabolise gluten. Most of the strains were classified within the phylum Firmicutes, Actinobacteria and Proteobacteria. The predominant genera were Lactobacillus, Staphylococcus, Clostridium, Bifidobacterium and Enterococcus (Table 2). No clear differences were associated with diagnosis, diet or genetic susceptibility. Isolated bacteria were characteristic of each volunteer and we did not find a pattern between groups. However, we found statistically significant differences in Lactobacillus. This genus was frequent in relatives and healthy volunteers but it was not found in samples from CD patients. We also detected a non-significant increase of Clostridium in CD patients.
Table 2 Cultivable bacteria involved in gluten metabolism isolated from faeces of healthy subjects, active coeliac disease patients and first-degree relatives (Number of samples with specific taxa and percentage of clones)
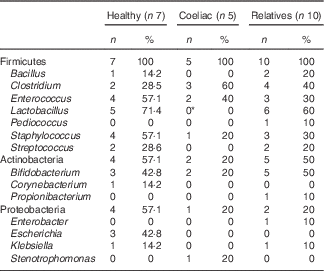
* Significant differences v. healthy volunteers and v. first-degree relatives (P<0·05).
On other hand, we studied the bacterial communities generated in liquid MCG-3 by PCR-DGGE. The PCR amplicon bands that were identified by sequencing are shown in Fig. 6. The bands corresponding to Bacteroides and Bifidobacterium were present in almost all the samples analysed. CD patients showed a significant increase in Clostridium compared with healthy volunteers and relatives as well as a reduction in the Lactobacillus group. A non-significant decrease of Bacteroides was also observed in healthy people.

Fig. 6 (a) Clustering of denaturing gradient gel electrophoresis (DGGE) profiles of bacteria growing in MCG-3 after incubation of faecal samples from healthy volunteers (H), untreated coeliac disease (CD) patients and first-degree relatives (R) of coeliac disease patients. The clusters are differentiated as A, B, C and D. (b) Table summarising the frequency of bacteria in DGGE bands of healthy volunteers, coeliac patients and relatives of coeliac patients. (c) Table showing the ability of faecal samples from healthy volunteers, coeliac patients and relatives of coeliac patients to hydrolyse gluten in MCG-3.
Richness of bacterial communities is associated with a higher hydrolysis against gluten
To analyse the relationships among the microbial communities, the UPGMA method based on the similarity indices of the DGGE profiles was carried out. The DGGE profiles of the PCR amplicons are indicated in Fig. 6. The DGGE profiles showed a main cluster D, which was mainly encompassed of relatives (eight out of ten) and CD patients (four out of five). Faecal samples from healthy volunteers were disperses (Fig. 1). Cluster D could be further distributed into three clusters: A, B and C. Interestingly, clusters A and B were dominated by communities of relatives and CD patients, respectively. We did not find differences in band profiles between samples from the same volunteer on GFD and normal diet (data not shown).
We also studied gluten hydrolysis by bacterial communities in MCG-3. In order to achieve this goal, we quantified the remaining gluten in MCG-3 after incubation with faeces using the ELISA kit G12. The faeces samples were classified into samples with low (0–40 %), medium (40–70 %) and high (70–100 %) hydrolytic efficacy towards gluten. We did not find a pattern between gluten consumption, CD diagnosis and the different clusters generated by the UPGMA method. Moreover, we did not find a correlation between bacteria (identified by DGGE and culture) and gluten hydrolysis. However, faecal samples with more bands showed a lower gluten concentration in media after incubation. Samples with a hydrolytic range between 70 and 100 % had an average of 6·9 bands, whereas low/medium hydrolysis had 5·1. The number of bands for each subject in every DGGE profile could be considered as an indicator of diversity.
Discussion
Although high-risk HLA alleles and exposure to gluten are frequent, most people with genetic susceptibility and gluten intake in their diet do not develop CD. Thus, other factors may be implicated in CD pathogenesis( Reference Camarca, Del Mastro and Gianfrani 21 – Reference Kagnoff 23 ). For a long time, it was considered that CD patients may have an abnormal gluten digestion due to impaired brush border peptidase activity in the small intestine, but this theory was abandoned( Reference Sjöström, Norén and Krasilnikoff 24 – Reference Bruce, Woodley and Swan 27 ). However, recently, the role of bacterial metabolism of gluten proteins in CD patients and healthy people has been raised. Altered bacterial metabolism could not only generate immunogenic gluten peptides but also lead to the production of pro-inflammatory substances such as sulphides, phenolic compounds and amines( Reference Davila, Blachier and Gotteland 9 , Reference Richardson, McKain and Wallace 28 ).
Dietary proteins are completely digested in the proximal small intestine( Reference Ganapathy, Gupta and Martindale 29 ). However, gluten proteins are unusually resistant to human digestive proteases generating high-weight oligopeptides such as 33-mer in the intestinal lumen capable of triggering the inflammatory process associated with CD( Reference Shan, Molberg and Parrot 7 ). In this study, we detected faecal peptidasic activity against 33-mer, the immunogenic peptide that contains six overlapping epitopes to which coeliacs react to. We also detected strong faecal proteolytic activity towards gluten proteins (FGA). Although the origin of these activities is still unknown, we hypothesised that these activities are mainly of bacterial origin. Dietary components not digested by human enzymes serve as substrates for intestinal bacteria; principally in the large intestine where a rich and complex microbiota resides( Reference Davila, Blachier and Gotteland 9 ). The genetic diversity of the intestinal microbiota produces different metabolic pathways that would not otherwise be present in the host( Reference Scott, Gratz and Sheridan 30 ). In fact, the human intestine is colonised by a heterogeneous proteolytic microbiota( Reference Richardson, McKain and Wallace 28 , Reference Macfarlane, Allison and Gibson 31 ), and faecal proteolytic activity is associated with compositional alterations in the intestinal microbiota( Reference Carroll, Ringel-Kulka and Ferrier 32 , Reference Midtvedt, Zabarovsky and Norin 33 ). Our group has recently isolated from the human gut thirty-six bacterial strains belonging to eleven different species with peptidasic activity against the 33-mer peptide. In addition, we described sixteen bacterial species with extracellular proteolytic activity against gluten proteins( Reference Caminero, Herrán and Nistal 14 ). Other bacteria through the digestive tract able to hydrolyse the 33-mer have been described( Reference Helmerhorst, Zamakhchari and Schuppan 34 – Reference Laparra and Sanz 36 ). In the present study, our results support the view that peptide fragments generated after the hydrolysis of 33-mer are characteristic of each volunteer, which correlates with the known fact that a microbiota fingerprint is characteristic of each individual( Reference Lozupone, Stombaugh and Gordon 37 ). Furthermore, we have shown that some bacterial communities generated in a liquid-enrichment culture hydrolyse gluten efficiently. Richness of the bacterial communities was also related to a higher hydrolysis potential against gluten. Thus, gluten peptides resistant to human digestive enzymes could be hydrolysed by a wide range of microbial proteases in the gastrointestinal tract.
Distinct associations between gut microbial communities and intestinal disorders have been identified, but no direct causal link to CD has been established( Reference Verdu 38 ). Several studies have shown differences in the composition of intestinal microbiota between CD patients and healthy people in the small and large intestines( Reference Nistal, Caminero and Vivas 2 – Reference Collado, Donat and Ribes-Koninckx 5 ). Although we did not find a strong link between bacteria involved in gluten metabolism and CD pathogenesis in this study, we assume that we have only studied a small portion of this population. Nevertheless, alterations in specific bacterial groups metabolising gluten such as Clostridium, Lactobacillus or Bacteroides were reported in CD patients in our study. These groups have been previously related to gluten metabolism and intestinal dysbiosis of CD patients. Moreover, it was previously shown that the SCFA and FTA, both indicators of bacterial metabolism, are altered in children with CD( Reference Tjellstrom, Stenhammar and Hogberg 39 ). We have observed that both SCFA and FTA values are increased in the adult population of CD patients, suggesting an alteration in the metabolic activity of the gut microbiota. Furthermore, our results show that the FGA is altered in CD patients with regard to healthy volunteers and first-degree relatives. These results are significant on GFD, suggesting a permanent alteration of the activity. CD patients on GFD showed a complete recovery in their intestinal mucosa. Therefore, an atrophic intestinal mucosa seems not to be the cause of these changes. Precisely, a previous study has shown that duodenal samples from untreated CD and treated CD patients have specific gliadinases that are absent in non-CD volunteers. The authors suggested the role of intestinal bacteria in these activities( Reference Bernardo, Garrote and Nadal 40 ). Thus, our results are consistent with the presence of intestinal functional dysbiosis in CD patients and a specific microbial activity that could lead to abnormal digestion of gluten proteins. It is possible that gluten intake generates immunogenic peptides or pro-inflammatory metabolites derived from an altered gluten metabolism in CD patients. Different bacterial genera could lead to an altered gut activity and chronic inflammation dependent on the HLA of the individual.
The fermentation of amino acids in the large intestine by bacterial metabolism produces mainly SCFA and nitrogenous compounds( Reference Davila, Blachier and Gotteland 9 ). Although SCFA are mostly produced by carbohydrates, protein compounds are also involved in SCFA production, being exclusive precursors of branched SCFA. In fact, it has been shown that high exogenous gluten intake in healthy people increases the concentration of faecal SCFA( Reference Caminero, Nistal and Arias 13 ). We compared the SCFA content in faeces based on CD diagnosis and genetic susceptibility and observed that SCFA was altered in faeces from CD patients( Reference Nistal, Caminero and Vivas 2 , Reference Tjellstrom, Stenhammar and Hogberg 39 ). These results may indicate not only the presence of colonic dysbiosis in CD patients but also a different amount of gluten peptides and amino acids in the colon as a consequence of abnormal digestion of proteins in the upper intestine by duodenal bacteria. First-degree relatives on ND showed a pattern of SCFA similar to CD patients and different from healthy people. Intestinal microbiota is highly influenced by genetic factors and shared bacteria could lead to a characteristic SCFA pattern. Although observed differences were seen in those on GFD, relatives showed higher branched SCFA than CD patients, which could be due to higher duration on GFD. Treated CD patients had decreased branched SCFA after at least 1 year on GFD and the microbiota capable to produce branched SCFA could be modified during this time. In contrast, first-degree relatives were only on GFD for 1 month. Thus, SCFA production was not different between CD patients and relatives, and the differences associated with the disease were linked to proteolytic patterns.
Dietary compounds not digested completely by human proteases and intestinal microbiota are eliminated in the faeces. Despite the proteolytic activity towards gluten peptides detected in this work, a part of gluten ingested through the diet reaches the faeces. We have shown that CD patients excreted less gluten in faeces than healthy people and first-degree relatives on ND. Previous studies have reported that gluten in faeces is influenced by gluten intake in the diet( Reference Caminero, Nistal and Arias 13 , Reference Comino, Real and Vivas 41 ). Although it is possible that CD patients avoided gluten-rich food because they suspected their clinical pathology or they had relative discomfort after gluten intake, other factors such as altered proteolytic activity or the increased intestinal permeability reported in CD patients could also be an influence. In fact, our results showed that FGA and FTA were increased in CD patients. Diagnostic groups with a low FGA such as healthy volunteers on normal diet showed a high amount of gluten in faeces, whereas CD patients with a high proteolytic activity excreted less amount of gluten. Furthermore, we showed that, despite a high-gluten-containing diet, volunteers with a high glutenasic activity excreted less gluten in faeces. FGA is directly related to 33-mer peptidasic activity and inversely to the amount of gluten in faeces; and gluten quantification was performed with ELISA, which detects a part of the 33-mer peptide. Therefore, the relationship between these parameters evidencing that gluten excretion is also influenced by intestinal proteolytic activity is clear. Gluten reduction in faeces from CD patients suggests a higher proteolytic activity in the intestine of these patients, which could be derived from an abnormal function of the intestinal microbiota.
In conclusion, results obtained for CD patients based on an increase in proteolytic activities and SCFA, as well as a lesser amount of gluten in faeces, suggest that gluten metabolism is altered in these patients.
Acknowledgement
The authors thank Dr Elena F. Verdú for critical revision of the manuscript.
This research was supported by grants from the Instituto de Salud Carlos III, Fondo de Investigación Sanitaria co-funded by FEDER (FIS PI10/02447); Junta de Castilla y León, Consejería de Sanidad (GRS 520/A/10). A. R. H. received a grant from the Junta de Castilla y León, co-funded by Fondo Social Europeo. J. P.-A. received a grant from the Ministerio de Educación, Cultura y Deporte del Gobierno de España.
A. C. F. and F. J. C. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of data analysis. Study concept and design: A. C. F., F. J. C., S. V. and J. M. G. R. d. M. Clinical selection and sampling: S. V. and L. V. A. Acquisition of data and analysis: A. C. F., E. N., A. R. H, J. P.-A., M. A. F. and S. M. A. Interpretation of data and drafting of the manuscript: A. C. F. and F. J. C. Critical revision of the manuscript: all the authors. Statistical analysis: A. C. F, L. V. A. and S. V.
There are no conflicts of interest.
Supplementary Material
For supplementary material/s referred to in this article, please visit http://dx.doi.org/10.1017/S0007114515002767













