In the dairy industry, it is a common practice to feed dairy cattle with a high concentrate diet in order to improve milk production. However, these diets can reduce ruminal pH and cause a shift in ruminal bacterial populations from cellulolytic-dominating (cellulose-digesting) species to amylolytic-dominating (starch-digesting) species of bacteria( Reference Metzler-Zebeli, Schmitz-Esser and Klevenhusen 1 ). Most of the known starch-digesting bacteria in the rumen are Gram-negative( Reference Bryant 2 , Reference Dehority and Dehority 3 ). Hence, the population changes caused by high grain feeding have been reported to associate with significant rises in the concentrations of lipopolysaccharide (LPS) in the rumen( Reference Khafipour, Li and Plaizier 4 ) and in acute-phase proteins, including LPS-binding protein, serum amyloid-A and haptoglobin, in peripheral blood( Reference Dehority and Dehority 3 ). Changes in the levels of LPS-binding protein are indicative of a systemic immune response that is caused by the translocation of LPS from the digestive tract( Reference Khafipour, Krause and Plaizier 5 ). In addition, some Gram-negative bacteria such as Escherichia coli are also responsible for many other diseases in ruminants, such as respiratory tract infections and mastitis( Reference Zimov, Botheras and Weiss 6 ). The clinical signs observed in Gram-negative bacterial infection are also mainly the result of the host's reaction to the bacterial components of cell walls, such as LPS( Reference Hoshino, Kawai and Sanjo 7 ).
The LPS are known to be a very potent antigen and, as a result, stimulate an intense host inflammatory response, and this inflammatory response can be induced experimentally by the administration of purified LPS( Reference Werling, Sutter and Arnold 8 ). It has been demonstrated that intravenous infusion of LPS reduced milk yield in dairy cattle, and the decreased animal performance may, in part, be affected by altered gastrointestinal digestibility, digesta passage rate or ruminal fermentation( Reference Steiger, Senn and Altreuther 9 , Reference Lohuis, Verheijden and Burvenich 10 ). Waggoner et al. ( Reference Waggoner, Loest and Turner 11 ) demonstrated that LPS infusion reduced passage rates of gastrointestinal solid and liquid contents. However, the effect of LPS treatment on ruminal fermentation has not been investigated. In addition, as the reduced passage rate of ruminal digesta after LPS treatment may probably lead to the accumulation of volatile fatty acids (VFA), this may result in a further drop in pH and an imbalanced microbial community in the rumen. However, there is little information available on the effect of LPS treatment on the changes in the ruminal microbiota. Recently, a study reported by Arthur et al. ( Reference Arthur, Perez-Chanona and Mühlbauer 12 ) showed that inflammation modifies gut microbial composition in colitis-susceptible IL-10-deficient mice. Given that the results from intestinal microbiota reported by Arthur et al. ( Reference Arthur, Perez-Chanona and Mühlbauer 12 ) also apply to the ruminal microbial communities, it is also reasonable to postulate that the immune activation by translocated LPS during high grain feeding could potentially disrupt the ruminal microbiota.
Thus, we hypothesised that LPS infusion into the circulating blood system could alter the ruminal microbiota and ruminal metabolism in dairy cattle. To test this hypothesis, a study was set up to characterise the effects of LPS challenge on the composition of ruminal microbial communities in dairy cattle. A secondary objective was to determine whether intravenous infusion of LPS would affect ruminal fermentation, degradability of forage and protein meal in the rumen, and gene expression related to VFA absorption in ruminal epithelium.
Materials and methods
All infusion and sampling procedures were approved by the Nanjing Agricultural University Institutional Animal Care and Use Committee before the experiment was initiated.
Animals and diets
A total of three primiparous lactating Holstein cows (474 (sd 50) kg body weight (BW); 15 (sd 1·3) kg/d DM intake (DMI), 260–285 d in lactation and 8·5 (sd 2·7) milk yield at the beginning of the trial) fitted with 10 cm ruminal cannulas (Anscitech) were used in a 3 × 3 Latin square design to assess the effects of three doses (0·0, 0·4 and 0·8 μg/kg BW) of bacterial LPS (E. coli 0111:B4; Sigma Chemical Company) on the composition of ruminal bacterial communities. The Latin square was balanced, in that each treatment followed a different treatment an equal number of times, to eliminate the bias of carry-over effects. The diets of the cows were formulated to meet or exceed the energy requirements (at 16 kg/d DMI) of Holstein cattle, yielding 12 kg milk/d with 3·50 % milk fat and 3·10 % true protein (see online supplementary Table S1). Diets were fed ad libitum as a total mixed ration to avoid the selection of dietary components. The cattle were fed at 07.00 and 18.00 hours (one-half of the allowed daily ration at each feeding). The experimental period was 16 d; the first 11 d were used for diet adaptation and the last 5 d were used for measurements. Throughout the experimental period, the cattle were housed in tie stalls and fed ad libitum to assure 5 % orts, and they were given free access to fresh water during the trial.
Lipopolysaccharide infusions
The infusion protocol used was modelled according to that of Werling et al. ( Reference Werling, Sutter and Arnold 8 ). The cattle were fitted with bilateral jugular vein catheters 24 h before the infusion began. The catheters were removed after the initiation of infusion (post-infusion hour). The treatments were dissolved in 100 ml of 0·9 % sterile saline and infused intravenously through the jugular catheter over a period of 100 min on day 12 of each experimental period. Infusions were administered via a Plum XL infusion pump with a 0·2 μm, low-protein-binding Sterile Acrodisc® 13 in-line filter. The LPS dose for each infusion day was assigned based on the average BW for the 3 d before infusion. Infusions began about 30 min after the morning daily milking (07.00 hours), and the cattle were offered their daily ration on experimental days at the initiation of infusion.
Consistent sampling collection and measurements
The feed offered and refused was weighed for five consecutive days on days 12, 13, 14, 15 and 16 of each experimental period to determine the DMI. On day 12 at 0 (before infusion), 2, 4, 8, 12, 24 and 48 h after LPS infusion, ruminal contents were collected from four locations in the rumen (ventral sac, reticulum and two from the feed mat in the dorsal rumen, approximately 100 g for each location). The ruminal samples were mixed and divided into two parts, and 50 g of the ruminal content samples were stored at − 80°C for DNA extraction. The remaining ruminal samples were filtered through four layers of cheesecloth; aliquots of the ruminal cheesecloth filtrates were immediately analysed for pH, and then centrifuged (20 000 g for 15 min at 4°C). The supernatant fluid was stored at − 20°C for analyses of VFA and NH3-N. VFA in the ruminal fluid were measured using a capillary column gas chromatograph (GC-14A; Shimadzu)( Reference Qin 13 ). NH3-N was measured by the indophenol method( Reference Weatherburn 14 ).
Samples of concentrates, maize silage, rice straw, Leymus chinensis and alfalfa hay were collected on days 12–14 of each period in the experiment. The samples were ground to pass through a 1 mm screen and analysed for DM( 15 ), ash( 15 ), diethyl ether extract( 15 ) and neutral-detergent fibre (NDF)( Reference Van Soest, Robertson and Lewis 16 ), NDF was determined using sodium sulphite and heat-stable amylase, and was inclusive of residual ash. Crude protein (N × 6·25) was determined by the method described by Krishnamoorthy et al. ( Reference Krishnamoorthy, Muscato and Sniffen 17 ).
Ruminal epithelial samples were collected via biopsy through ruminal fistulae on day 12 at 4 h after LPS infusion. In brief, two biopsy samples (approximately 150 mg papillae) were taken (approximately 2–8 cm apart) from each cow. The biopsy samples were washed twenty times in ice-cold PBS, and the samples were fixed in RNAlater (Life Technologies) RNA stabilisation solution, according to the manufacturer's instructions, and stored at − 80°C until RNA isolation and analysis.
Ruminal disappearance study
NDF, protein and organic matter (OM) disappearances in the rumen were estimated for alfalfa and soyabean meal samples using the nylon bag technique on day 12 of each experimental period at 0 (before infusion), 2, 4, 8, 12, 24, 48 and 72 h after LPS infusion( Reference Ørskov and McDonald 18 ). The samples of the soyabean meal and alfalfa were oven-dried at 60°C for 48 h, and milled to a 1 mm screen (Polymix® PX-MFC; Kinematica AG). The bags (7 × 14 cm) made from Dacron cloth with a pore size of 38 μm were each filled with approximately 5 g of dried (60°C) feed samples. The bags were tightly sealed. All samples were prepared in duplicate at each time point, and a blank bag containing no sample was also prepared for each removal time. All bags were incubated simultaneously in the rumen of the fistula animal on day 12 of each experimental period at 0 h (before infusion). The bags for each feed sample and blank were removed after 0, 2, 4, 8, 12, 24, 48 and 72 h incubation. Immediately after the bags were removed from the rumen, they were washed with cold tap water until clear and dried in a forced-air oven at 60°C for 72 h. The bags were weighed and the residues were removed and then analysed for NDF and OM in alfalfa and protein and for OM in the soyabean meal. All bag feed samples were collected for their corresponding blank. The bags were weighed and tested according to the procedure described by Ørskov & McDonald( Reference Ørskov and McDonald 18 ). All the samples were analysed for DM, ash and crude protein using the procedure of the AOAC( 15 ), and NDF was analysed according to Van Soest et al. ( Reference Van Soest, Robertson and Lewis 16 ).
DNA isolation
The ruminal samples collected at 0, 2, 8, 24 and 48 h were used for analyses of microbial profiles only. In brief, 1 ml of ruminal content was used for DNA extraction. DNA was extracted by a bead-beating method using a mini-bead beater (Biospec Products), followed by phenol–chloroform extraction( Reference Frey, Pell and Berthiaume 19 ). The solution was precipitated with ethanol and the pellets were suspended in 50 μl-Tris–EDTA buffer. DNA was quantified using a Nanodrop spectrophotometer (Nyxor Biotech) following staining using a Quant-it Pico Green dsDNA kit (Invitrogen Limited). DNA samples were stored at − 80°C until further processing.
DNA pyrosequencing
The following universal primers were applied for the amplification of the V3–V6 region of the 16S rRNA gene: forward primer, 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGNNNNNN ACTCCTACGGGAGGCAGCAG-3′ (the italicised sequence is the 454 Life Sciences primer A and the bold sequence is the broadly conserved bacterial primer 338F; NNNNNN designates the sample-specific six-base barcode used to tag each PCR product); reverse primer, 5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG CRRCACGAGCTGACGAC-3′ (the italicised sequence is the 454 Life Sciences primer B and the bold sequence is the broadly conserved bacterial primer 1061R). The cycling parameters were as follows: 5 min initial denaturation at 95°C; twenty-five cycles of denaturation at 95°C (30 s), annealing at 55°C (30 s), elongation at 72°C (30 s); final extension at 72°C for 5 min. For pyrosequencing, three separate PCR of each sample were pooled. The PCR products were separated by 1 % agarose gel electrophoresis and purified by using a QIAquick Gel extraction kit (Qiagen). Amplicons were quantified using a Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen). Equal concentrations of amplicons were pooled from each sample. The amplicons were sequenced as recommended by the manufacturer's instructions. The end fragments were blunted and tagged on both ends with one of forty-five ligation adaptors, which contained a unique 10 bp sequence and a short four-nucleotide sequence (TCAG) called the sequencing key; these were recognised by the system software and the priming sequences.
Pyrosequencing data analysis
The sequences were processed using the program MOTHUR version 1.29.0( Reference Schloss, Westcott and Ryabin 20 ). 16S rRNA reads were decoded based on the 5 bp sample-specific barcodes and processed to remove poor-quality sequences. To reduce sequencing errors, the shhh.flows command was applied, which is the MOTHUR implementation of the AmpliconNoise algorithm( Reference Quince, Lanzen and Davenport 21 ). Quality filters were applied to trim and remove the sequences as follows: sequences less than 200 bp in length; average quality score less than 35; homopolymers longer than eight nucleotides and more than two different bases to the primer. In order to obtain a non-redundant set of sequences, unique sequences were determined and used to align against the SILVA reference alignment database( Reference Pruesse, Quast and Knittel 22 ); chimeras were removed using chimera.uchime (http://drive5.com/uchime); sequences identified as being of eukaryotic origin were removed; and the candidate sequences were screened and pre-clustered to eliminate outliers; a distance matrix was generated from the resulting sequences. Sequences were clustered into operational taxonomic units (OTU) using the furthest-neighbour algorithm. Representative sequences from OTU at a 0·03 distance were obtained and classified using the Ribosomal Database Project's Bayesian classifier( Reference Wang, Garrity and Tiedje 23 ). Rarefaction curves and Good's coverage were calculated to quantify the coverage and sampling effort. Community diversity was estimated using the ACE (abundance-based coverage estimator), Chao1 and Shannon–Wiener (Shannon) indices. The unweighted Unifrac distance method( Reference Lozupone and Knight 24 ) was used to perform a principal coordinate analysis (PCoA), and a distance-based AMOVA (analysis of molecular variance) was conducted to assess significant differences between the samples. A double hierarchical analysis was conducted using the unweighted pair-group method with arithmetic mean and Manhattan distance with no scaling in the Number Cruncher Statistical System (NCSS 2007) software (NCSS).
Extraction of ruminal tissue RNA and reverse transcription
Total RNA was extracted from ground ruminal tissue using TRIzol (Invitrogen) as described by Wang et al. ( Reference Wang, Akers and Jiang 25 ). RNA concentration was determined by measuring absorbance at 260 and 280 nm using NanoDrop (ND-1000; NanoDrop Technologies). All samples had an absorbance ratio (260:280) between 1·90 and 2·13, indicating high RNA purity. The samples were then diluted to contain 100 ng RNA/μl. All RNA samples were treated with DNase I (Invitrogen) to remove potential genomic DNA contamination. The Superscript II kit (Invitrogen) was used to synthesise single-strand complementary DNA.
Quantitative real-time PCR
Primers were selected to amplify the genes encoding for three monocarboxylic acid transporters (MCT1, MCT2 and MCT4) involved in the absorption of VFA( Reference Naeem, Drackley and Stamey 26 ) and Na+/K+ transporting α-1 polypeptide (Na+/K+-ATPase)( Reference Kuzinski, Zitnan and Viergutz 27 ). The primer information including sequences is shown in online supplementary Table S2. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene( Reference Wang, Akers and Jiang 25 ). All primers were synthesised by Invitrogen Life Technologies. Real-time quantitative PCR of target genes and GAPDH were performed using the ABI 7300 real-time PCR system (Applied Biosystems) with fluorescence detection of SYBR green dye. Amplification conditions were as follows: 95°C for 30 s, followed by forty cycles composed of 5 s at 95°C and 31 s at 57·5°C (for GAPDH) or 60°C (for others). Each sample contained 1–10 ng complementary DNA in 2 × SYBR Green PCR Master Mix (Takara Bio) and 200 nmol/l of each primer in a final volume of 20 μl. All measurements were performed in triplicate. A reverse-transcription-negative blank of each sample and a no-template blank served as negative controls. The relative amount of each studied mRNA was normalised to GAPDH mRNA levels as a housekeeping gene, and data were analysed according to the 2− ΔΔC T method. The primers and amplicon sizes of all genes are presented in online supplementary Table S2.
Data analysis
Ruminal pH, VFA, degradation disappearance and microbial data were analysed with the MIXED procedure of SPSS (version 16; SPSS, Inc.) according to the following model:
where Y ijk is the ith observation (ruminal pH, NH3-N, specific VFA concentration (mmol/l) and microbial data) from the jth cattle; μ is the overall mean; D i is the fixed effect of LPS (i= 1–3 for dosage); G j is the random cattle effect (j= 1–3); T k is the fixed effect of period (k= 1–3 for periods); TD ik is the fixed effect of the LPS treatment × period interaction; e ijk is the residual error for the kth observation from the jth cattle; residual terms are assumed to follow normal distributions. Measurements collected at different times from the same cattle were considered as repeated measures in the ANOVA. The sums of squares were further partitioned by orthogonal polynomial contrasts to study the linear and quadratic effects of the treatment. All P values from multiple comparison tests for microbial data were adjusted by the false discovery rate. Significance was declared at P≤ 0·05.
Gene expression data were analysed using the generalised linear model procedure of SPSS (version 16; SPSS, Inc.) according to the following model:
where Y ijk is the ith observation (MCT1, MCT2 and MCT4 or Na+/K+-ATPase data) from the jth cattle; μ is the overall mean; D i is the fixed effect of LPS (i= 1–3 for dosage); G j is the random cattle effect (j= 1–3); T k is the fixed effect of period (k= 1–3 for periods); TD ik is the fixed effect of the LPS treatment × period interaction; e ijk is the residual error for the kth observation from the jth cattle; residual terms are assumed to follow normal distributions. Orthogonal polynomial contrasts were used to test for the linear and quadratic effects of LPS treatment. Significance was declared at P≤ 0·05.
Results
Feed intake and rectal temperatures
Figure 1 shows the time course of changes in DMI and rectal temperatures. Compared with the control, DMI was not affected (P= 0·156) by the LPS challenge (Fig. 1(a)). Rectal temperatures in cattle challenged with LPS increased relative to that in the saline-infused cattle (linear effect, P< 0·001; quadratic effect, P< 0·001; Fig. 1(b)). During the 3 h period of fever peak, the average maximum rectal temperature for the cattle was 39·62 ± 0·58°C during the 0·8 μg/kg BW LPS treatment day v. 38·60 ± 0·10°C for the control day.
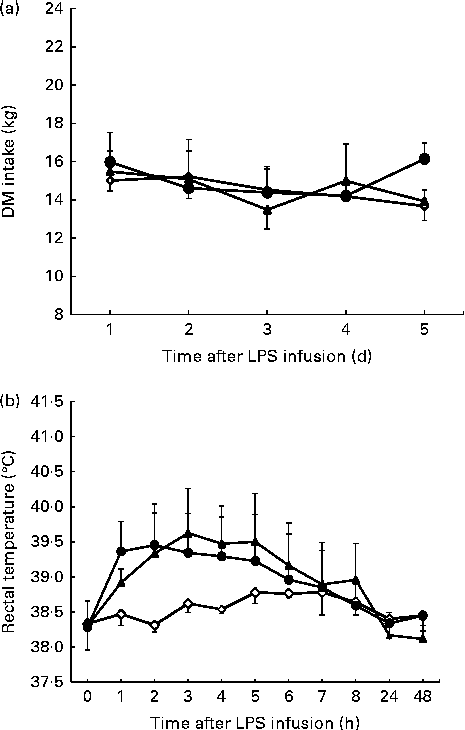
Fig. 1 DM intake (a) and rectal temperature (b) following lipopolysaccharide (LPS) challenge in dairy cattle. Values are means (n 3), with their standard errors represented by vertical bars. ![]() , 0 μg LPS/kg body weight;
, 0 μg LPS/kg body weight; ![]() ; 0·4 μg LPS/kg body weight;
; 0·4 μg LPS/kg body weight; ![]() , 0·8 μg LPS/kg body weight.
, 0·8 μg LPS/kg body weight.
Ruminal pH, ammonia nitrogen and volatile fatty acid concentrations
Ruminal pH decreased linearly (P< 0·001) following intravenous LPS administration (Fig. 2(a)). There were linear (P< 0·001) increases in the concentrations of acetate (Fig. 2(b)), propionate (Fig. 2(c)), butyrate (Fig. 2(d)), total branched-chain VFA (Fig. 2(e)), total VFA (Fig.2 (f)) and NH3-N (Fig. 2 (h)) following the LPS challenge. LPS administration linearly increased (P= 0·006) the ratio of acetate:propionate (Fig. 2(g)).

Fig. 2 Ruminal pH (a), volatile fatty acid (VFA) (b–f) and NH3-N (h) concentrations, and acetate:propionate ratio (g) in the ruminal fluid of dairy cattle challenged with lipopolysaccharide (LPS). Values are means (n 3), with their standard errors represented by vertical bars. ![]() , 0 μg LPS/kg body weight;
, 0 μg LPS/kg body weight; ![]() ; 0·4 μg LPS/kg body weight;
; 0·4 μg LPS/kg body weight; ![]() , 0·8 μg LPS/kg body weight.
, 0·8 μg LPS/kg body weight.
Neutral-detergent fibre, protein and organic matter degradability of feeds
Ruminal NDF, protein and OM disappearances for the soyabean meal and alfalfa are shown in Fig. 3. LPS infusion linearly decreased (P< 0·001) the NDF degradability of alfalfa (Fig. 3(a)), the OM degradability of alfalfa (Fig. 3(b)) and soyabean meal (Fig. 3(c)) and the protein degradability of the soyabean meal (Fig. 3(d)).

Fig. 3 Ruminal digestibility of alfalfa (a, b) and soyabean meal (c, d) in dairy cattle challenged with lipopolysaccharide (LPS). Values are means (n 3), with their standard errors represented by vertical bars. ![]() , 0 μg LPS/kg body weight;
, 0 μg LPS/kg body weight; ![]() ; 0·4 μg LPS/kg body weight;
; 0·4 μg LPS/kg body weight; ![]() , 0·8 μg LPS/kg body weight. NDF, neutral-detergent fibre; OM, organic matter.
, 0·8 μg LPS/kg body weight. NDF, neutral-detergent fibre; OM, organic matter.
Gene expression
Among the SCFA transporters, MCT1, MCT2 and MCT4 (Fig. 4) were evaluated for their gene expression. There was no effect (P>0·10) of the LPS challenge on the expression of any of the genes evaluated for the transport of VFA. Additionally, LPS challenge did not affect (P>0·10) the relative expression of the Na + /K +-ATPase gene (Fig. 4).

Fig. 4 Quantitative real-time PCR analysis of the expression levels of Na
+
/K
+-ATPase, monocarboxylic acid transporter-1 (MCT1), MCT2 and MCT4 in the ruminal epithelium of dairy cattle challenged with lipopolysaccharide (LPS). Values are means (n 3), with their standard errors represented by vertical bars. ![]() , 0 μg LPS/kg body weight;
, 0 μg LPS/kg body weight; ![]() ; 0·4 μg LPS/kg body weight;
; 0·4 μg LPS/kg body weight; ![]() , 0·8 μg LPS/kg body weight.
, 0·8 μg LPS/kg body weight.
General DNA sequencing observations
The deep sequencing of forty-five ruminal samples from the cattle provided a detailed view of the ruminal microbiome of the cattle. A total of 296 606 sequences were derived for bacterial diversity analysis; of a total of 236, 375 remained post-filtering including chimera and low-confidence singleton removal, and 5040 OTU were identified at the 97 % similarity level. The sequence number per sample ranged from 4200 sequences (P3-B3-D8-T2) to 8496 sequences (P1-B3-D0-T4) with a mean of 5252 (sd 697) (n 45) sequences per sample (see online supplementary Table S3). A total of eighteen phyla were observed to be distributed among all cattle on all diets (see online supplementary Fig. S1). These are listed in order of average abundance with their respective ranges as follows: Firmicutes (59·3 %, 42·4–75·6 %); Bacteroidetes (34·3 %, 19·3–50·3 %); unclassified Bacteria (2·6 %, 1·1–4·4 %); Tenericutes (1·3 %, 0·4–2·6 %); Actinobacteria (1·2 %, 0·5–2·6 %); Spirochaetes (0·48 %, 0·07–1·44 %); Proteobacteria (0·47 %, 0·05–2·86 %); Cyanobacteria (0·13 %, 0·02–0·42 %); Lentisphaerae (0·11 %, 0–0·38 %); Fibrobacteres (0·06 %, 0–0·23 %); Chloroflexi (0·035 %, 0–0·12 %); Elusimicrobia (0·034 %, 0–0·18 %); Synergistetes (0·016 %, 0–0·08 %); Chlorobi (0·011 %, 0–0·1 %); Verrucomicrobia (0·004 %, 0·000–0·04 %); Armatimonadetes (0·003 %, 0–0·04 %); Planctomycetes (0·001 %, 0–0·03 %); Fusobacteria (0·001 %, 0–0·02 %). Greater than 99·9 % of total bacterial abundance was observed in the first ten phyla.
At the genus level, the sequences were assigned to 205 different genera. Of these, the control, 0·4 μg/kg BW and 0·8 μg/kg BW groups were represented by 165, 151 and 136 genera, respectively (Table 1). The average numbers of OTU returned for each diet were as follows: control, 3450; 0·4 μg LPS/kg BW group, 3262; 0·8 μg LPS/kg BW group, 3265 (data not shown).
Table 1 Number of sequences and number of bacterial genera identified to be common to each group at the genus level

BW, body weight.
* The total number of genera identified in each group, respectively.
† The number of genera that are only detected across the entire sample collection in the control or lipopolysaccharide treatment.
Diversity of ruminal bacterial communities
Rarefaction was performed at the OTU level, and rarefaction curves demonstrated that (1) diversity differed between individual samples (number of OTU observed) and (2) the majority of bacterial diversity (OTU) was not uncovered, as many of the plots were not close to becoming asymptotic (see online supplementary Fig. S2). Alpha diversity measures (Shannon index, ACE and Chao1) are presented for each host species in Table 2. Alpha diversity measures indicated that LPS challenge did not seem to affect biodiversity indices, such as ACE, Chao1 and Shannon indices, in the rumen.
Table 2 Summary overview of estimated operational taxonomic units per diet through abundance-based coverage estimator (ACE), Chao1 and Shannon indices averaged across the animals per treatment (n 3)

LPS, lipopolysaccharide; BW, body weight.
A PCoA of overall diversity at the 3 % cut-off OTU level, based on an unweighted UniFrac metric, was also performed to compare all samples. This analysis showed that the control and LPS-treated communities were highly significantly different from each other (AMOVA, P< 0·05). Comparison of bacterial communities by PCoA showed that cattle infused with 8 μg LPS/ml were separated from those receiving the control and 0·4 μg/kg treatment (Fig. 5). In general, PCoA with unweighted UniFrac values showed that principal coordinate 1 was 26·6 % of the variation and principal coordinate 2 was 8·06 % of the variation.

Fig. 5 Principal coordinate (PC) analysis (PCoA) results showing the relationships of ruminal bacterial communities of dairy cattle challenged with different levels of lipopolysaccharide (LPS). The PCoA plots were constructed using the unweighted UniFrac method at the 3 % cut-off operational taxonomic unit level. ![]() , 0 μg LPS/kg body weight;
, 0 μg LPS/kg body weight; ![]() ; 0·4 μg LPS/kg body weight;
; 0·4 μg LPS/kg body weight; ![]() , 0·8 μg LPS/kg body weight.
, 0·8 μg LPS/kg body weight.
Influence of lipopolysaccharide treatment on ruminal microbiota at the levels of phyla, genera and operational taxonomic units
The changes of microbial abundance at the phylum level after LPS treatment are presented in Fig. 6. LPS infusion linearly increased the abundance of Firmicutes (P< 0·001), and linearly decreased the percentage of Bacteroidetes (P< 0·001), Tenericutes (P= 0·001), Spirochaetes (P= 0·041), Chlorobi (P= 0·014) and Lentisphaerae (P= 0·027).

Fig. 6 Influence of lipopolysaccharide (LPS) challenge on the ruminal microbiota of dairy cattle at the phylum level. (Only the percentage of phyla that were significantly affected (P< 0·05) by lipopolysaccharide challenge are presented.) ![]() , Chlorobi;
, Chlorobi; ![]() , Lentisphaerae;
, Lentisphaerae; ![]() , Spirochaetes;
, Spirochaetes; ![]() , Tenericutes;
, Tenericutes; ![]() , unclassified Bacteria;
, unclassified Bacteria; ![]() , Bacteroidetes;
, Bacteroidetes; ![]() , Firmicutes. BW, body weight.
, Firmicutes. BW, body weight.
The influence of the LPS challenge on the ruminal microbiome was also observed from the double hierarchical cluster analysis on the top fifty most abundant genera ( ≥ 99 % of the total number of bacterial genera observed), which were clustered by LPS treatment (see online supplementary Fig. S3). Among these genera, unclassified Ruminococcaceae, unclassified Christensenellaceae, unclassified Bacteroidales, unclassified Rikenellaceae, unclassified Lachnospiraceae and Prevotella occurred together in one cluster, while other forty-four genera co-habited in another cluster. The changes in microbial abundance at the genus level after LPS treatment (P< 0·05) are presented in Fig. 7 . In general, when compared with the control group, LPS infusion linearly decreased the percentage of Prevotella (P< 0·001), Anaeroplasma (P< 0·001), Victivallis (P= 0·019), Saccharopolyspora (P= 0·029), Microbacterium (P= 0·028), unclassified Bacteroidales (P= 0·001), unclassified Rikenellaceae (P= 0·04), unclassified Prevotellaceae (P= 0·012), unclassified Mollicutes (P= 0·029) and unclassified Chlorobiales (P= 0·002), and linearly increased the percentage of Atopobium (P= 0·041), Anaerotruncus (P= 0·003), Syntrophococcus (P= 0·038), Succinivibrio (P= 0·023), unclassified Christensenellaceae (P< 0·001), unclassified Lachnospiraceae (P= 0·025) and unclassified Clostridiales (P= 0·008). As LPS infusion increased, there was a quadratic effect on the abundance of Pyramidobacter (P= 0·015), Anaerobiospirillum (P= 0·021), Paraprevotella (P= 0·011), unclassified Desulfuromonadales (P= 0·005), unclassified Erysipelotrichaceae (P= 0·036) and unclassified Bacteria (P= 0·032).

Fig. 7 Influence of lipopolysaccharide (LPS) challenge on the ruminal bacterial microbiota of dairy cattle at the genus level. (Only the percentage of genera that were significantly affected (P< 0·05) by lipopolysaccharide challenge are presented.) ![]() , Succinivibrio;
, Succinivibrio; ![]() , Anaerobiospirillum;
, Anaerobiospirillum; ![]() , Pyramidobacter;
, Pyramidobacter; ![]() , Microbacterium;
, Microbacterium; ![]() , unclassified Chlorobiales;
, unclassified Chlorobiales; ![]() , Victivallis;
, Victivallis; ![]() , Paraprevotella;
, Paraprevotella; ![]() , unclassified Desulfuromonadales;
, unclassified Desulfuromonadales; ![]() , Syntrophococcus;
, Syntrophococcus; ![]() , Anaerotruncus;
, Anaerotruncus; ![]() , Spirochaeta;
, Spirochaeta; ![]() , Atopobium;
, Atopobium; ![]() , Anaeroplasma;
, Anaeroplasma; ![]() , unclassified Erysipelotrichaceae;
, unclassified Erysipelotrichaceae; ![]() , unclassified Mollicutes;
, unclassified Mollicutes; ![]() , unclassified Bacteria;
, unclassified Bacteria; ![]() , unclassified Prevotellaceae;
, unclassified Prevotellaceae; ![]() , unclassified Clostridiales;
, unclassified Clostridiales; ![]() , unclassified Lachnospiraceae;
, unclassified Lachnospiraceae; ![]() , unclassified Rikenellaceae;
, unclassified Rikenellaceae; ![]() , unclassified Bacteroidales;
, unclassified Bacteroidales; ![]() , unclassified Christensenellaceae;
, unclassified Christensenellaceae; ![]() , Prevotella. BW, body weight.
, Prevotella. BW, body weight.
Discussion
Effect of lipopolysaccharide infusion on rectal temperature, ruminal fermentation, DM intake, diet digestibility and gene expression of Na+/K+-ATPase and monocarboxylic acid transporter-1, -2 and -4
The jugular vein infusion of LPS is an established experimental model used to investigate the local and systemic inflammatory responses of lactating dairy cattle( Reference Jain, Schalm and Lasmanis 28 ). Werling et al. ( Reference Werling, Sutter and Arnold 8 ) asserted that a continuous infusion more closely mimics the effects of a true bacterial infection. In the literature, doses of 0·025( Reference Borderas, De Passillé and Rushen 29 ), 0·01( Reference Jacobsen, Andersen and Toelboell 30 ), 0·05( Reference Borderas, De Passillé and Rushen 29 ), 0·1( Reference Jacobsen, Andersen and Toelboell 30 , Reference Elsasser, Kahl and Rumsey 31 ), 0·2( Reference Kahl, Elsasser and Blum 32 ), 0·5( Reference Waldron, Nishida and Nonnecke 33 ), 0·6( Reference Sartin, Elsasser and Kahl 34 ), 1·0( Reference Elsasser, Kahl and Rumsey 31 ), 1·5( Reference Waldron, Kulick and Bell 35 ) and 2·0( Reference Steiger, Senn and Altreuther 9 , Reference Waldron, Kulick and Bell 35 ) μg LPS/kg BW have been used in cattle. In the present study, the doses of LPS used (0–0·8 μg/kg BW) were relatively low, and the results agreed with previous reports that LPS challenge caused increased rectal temperature( Reference Shuster, Harmon and Jackson 36 , Reference Perkins, VandeHaar and Burton 37 ); however, the feed intake was not affected by the infusion of LPS. This result is not consistent with the reports of Steiger et al. ( Reference Steiger, Senn and Altreuther 9 ) (2 μg LPS/kg BW) and Waggoner et al. ( Reference Waggoner, Loest and Turner 11 ) (2 μg LPS/kg BW), who both reported that the LPS challenge decreased the feed intake in cattle. The lack of an effect on feed intake may be due to the mild doses used. Johnson & von Borell( Reference Johnson and von Borell 38 ) reported that the reduction of food intake by LPS administration was dose-dependent and short-lived in swine. Borderas et al. ( Reference Borderas, De Passillé and Rushen 29 ) reported that a low dose of 0·025 or 0·05 μg/kg BW of LPS injection did not affect the feed intake in dairy calves. In general, those results showed that low dosage of LPS challenge might not affect the feed intake, suggesting that moderate sick animals can still consume enough feed to support all body functions of the host animal.
In the present study, concentrations of ruminal VFA were generally greater in LPS-treated cattle, which was indicative of either increased OM degradability and feed intake or decreased VFA absorption and passage rate in the ruminal fluid through the reticulum–omasum orifice( Reference Peters, Shen and Chester 39 ). The results of digestive efficiency in the present study revealed that LPS infusion decreased the OM degradability of the soyabean meal and alfalfa, but did not affect DMI. Therefore, the higher ruminal VFA concentrations in the LPS-treated cattle were possibly related to the decrease in VFA absorption or the passage of ruminal contents. However, little information was available on whether VFA absorption through the ruminal epithelium can be affected by LPS challenge in cattle. Nevertheless, Albin et al. ( Reference Albin, Wubben and Rowlett 40 ) reported that nutrient uptake of female Meishan pigs and female Yorkshire pigs responded in opposite ways to LPS challenge. Meishan pigs responded by increasing nutrient uptake, whereas Yorkshire pigs responded by decreasing nutrient uptake after a 4 h exposure to LPS. In the present study, to investigate whether LPS challenge could affect VFA absorption in the rumen, we evaluated the effect of LPS treatment on the gene expression of the Na + /K + -ATPase, MCT1, MCT2 and MCT4 mRNA transcripts. Among the four genes, Na + /K + -ATPase is a key transport element required for the establishment of electrochemical gradients that drive cellular transport and substrate flow across epithelia( Reference Zouzoulas, Dunham and Blostein 41 ). In ruminal epithelium, the Na+-motive force generated by the Na+/K+-ATPase is essential for the absorption of Na and is indirectly related to short fatty acid transport via the Na+/H+ exchanger( Reference Harrison, Keynes and Rankin 42 ). However, in the present study, the gene expression of Na + /K + -ATPase was not affected by the LPS challenge, which indicated that there was no apparent relationship between the accumulation of ruminal VFA and the gene expression of Na + /K + -ATPase during LPS treatment.
In the rumen, the uptake of VFA by epithelial cells lining the alimentary canal is partially dependent on carrier-mediated transporters, such as MCT1, MCT2 and MCT4( Reference Naeem, Drackley and Stamey 26 ), in addition to passive diffusion across the apical membrane of epithelial cells( Reference Sehested, Diernaes and Moller 43 ). Previous studies revealed that gene expressions of MCT1, MCT2 and MCT4 were modulated by ruminal VFA concentration and diet energy concentration( Reference Naeem, Drackley and Stamey 26 , Reference Borthakur, Saksena and Gill 44 ). In human colon epithelial cells, the SCFA butyrate has been shown to stimulate the promoter activity of MCT1 and to induce the up-regulation of MCT1 at the mRNA (5·7-fold) and protein (5·2-fold) levels( Reference Cuff, Lambert and Shirazi-Beechey 45 ). Using the ruminal epithelial cells obtained from sheep fed a mixed hay/concentrate diet, Kuzinski et al. ( Reference Kuzinski, Zitnan and Viergutz 27 ) reported that the protein and mRNA expression of MCT1 in ruminal epithelial cells were increased compared with the animals fed the hay diet. Similarly, Metzler-Zebeli & Zebeli( Reference Metzler-Zebeli and Zebeli 46 ) reported that the ruminal epithelium responds to a 60 % grain diet with an up-regulation of MCT1 expression, but with a down-regulation of MCT4 expression compared with 0 and 30 % grain diets in goats. In the present study, although total VFA concentrations were increased by the intravenous LPS challenge, no significant changes were observed in the gene expression of MCT1, MCT2 and MCT4 between the control and LPS challenge groups. This difference may be explained by the complex modulation of host gene expression. Indeed, previous studies revealed that besides substrate (butyrate) -induced up-regulation of MCT1 expression, MCT1 and its chaperon CD147 in intestinal epithelial cells can be down-regulated by the increase in the levels of inflammation cytokines such as interferon-γ and TNF-α in mice( Reference König, Fischer and Schlotte 47 , Reference Thibault, De Coppet and Daly 48 ), and this indicates that the positive effect of butyrate on the expression of MCT1 in mice could be weakened by inflammation. Therefore, in the ruminant, we postulate that although the increased VFA concentration in the rumen could up-regulate MCT1 expression, this regulation could be weakened by the systemic LPS challenge, consequently leading to no significant change in MCT1 expression. This may partly explain the facts in the present study that no significant differences were observed in the expression of MCT1, MCT2 and MCT4 between the control and LPS treatment groups. These findings suggest that the increased VFA concentration during LPS challenge may not be related to ruminal wall absorption. It is possible that the accumulation of ruminal VFA during LPS challenge may be linked to the slower passage of the ruminal fluid through the reticulum–omasum orifice, as suggested by Lohuis et al. ( Reference Lohuis, Verheijden and Burvenich 10 ) and Waggoner et al. ( Reference Waggoner, Loest and Turner 11 ), who reported that LPS infusion decreased the passage rate of gastrointestinal contents and ruminal motility, and the mechanisms responsible for the alternation in the passage rate involved in raised body temperature and smooth muscle relaxation in the gastric wall that were induced by PG after LPS challenge( Reference Lohuis, Verheijden and Burvenich 10 ).
It is well known that acute and chronic acidosis, which often occurs with gastrointestinal endotoxin translocation, are prominent problems for ruminants associated with the management practice of high grain feeding( Reference Khafipour, Krause and Plaizier 5 ). Borderas et al. ( Reference Borderas, De Passillé and Rushen 29 ) reported that LPS injections (0·025 or 0·05 μg/kg BW) into the jugular vein reduced the time spent ruminating in dairy calves, and this indicated that LPS infusion could reduce rumination, and resulted in a decrease in saliva production and rumen buffering. In the present study, LPS challenge also led to the greater accumulation of total VFA in the rumen. Thus, the decrease in rumen buffering, combined with a greater accumulation of total VFA in the rumen during LPS challenge could result in a dramatic drop in ruminal pH. These findings indicate that translocated LPS during subacute ruminal acidosis may aggravate ruminal acidosis and lead to a double detriment to rumen health. These results also suggest that, in addition to feed management practices, e.g. sodium bicarbonate as a dietary buffer and monensin and lasalocid as additives, anti-endotoxin therapy should be used to minimise undesirable changes in ruminal fermentation characteristics during ruminal acidosis.
Ruminal pH, together with the microbial population, the nature of substrates, environmental factors such as temperature and existence of cations, have been suggested as factors governing the metabolism of ruminal micro-organisms( Reference Metzler-Zebeli, Schmitz-Esser and Klevenhusen 1 , Reference Khafipour, Li and Plaizier 4 ). In the present study, LPS challenge resulted in a decrease in the degradability of protein, NDF and OM in the rumen. These results may be partly explained by the lower ruminal pH in the LPS-treated group, because low ruminal pH can negatively affect microbial metabolism and nutrient degradation( Reference Khafipour, Li and Plaizier 4 ). These results also indicate that LPS infusion disrupted ruminal metabolism. In general, these findings demonstrated that the intravenous infusion of an endotoxin alters ruminal environment and ruminal metabolism in cattle.
Effect of lipopolysaccharide infusion on ruminal bacterial microbiota
The present study characterises temporal changes in the ruminal microbiota of dairy cows in response to LPS challenge. In line with other previous studies( Reference Jami and Mizrahi 49 , Reference Pitta, Pinchak and Dowd 50 ), Bacteroidetes and Firmicutes in the control group constituted a major fraction of the total sequencing reads, which totally accounted for up to 93·6 % of all 16S rRNA gene sequences (see online supplementary Fig. S1). The predominant genera in the control group (>1 % of the total sequence) were Prevotella, Butyrivibrio, Succiniclasticum, Ruminococcus and Acetitomaculum, and the members of Prevotella were the most abundant of all genera identified; this finding is in agreement with earlier studies employing different methods, including analysis of 16S rRNA gene clone libraries( Reference Bekele, Koike and Kobayashi 51 ), 16S rDNA pyrosequencing( Reference Jami and Mizrahi 49 , Reference Pitta, Pinchak and Dowd 50 ) and quantitative PCR analysis( Reference Stevenson and Weimer 52 ).
In the present study, despite no significant differences in the numbers of alpha diversity measures between the control and LPS treatment samples, the numerical decrease in the value of the Shannon index, Chao1 or Ace were observed in the 0·8 μg LPS/kg BW group compared with the control group. In addition, a PCoA plot based on unweighted UniFrac values revealed strong clustering of the control and LPS treatment samples, indicating that the structure of ruminal bacterial communities in the control sample was distinct from that of the ruminal microbiota after LPS challenge (Fig. 5). In addition, AMOVA showed differentiation among the populations, which indicated that ruminal microbial populations differed greatly. Therefore, the present study demonstrated the hypothesis that LPS infusion altered the structure and composition of ruminal bacterial communities. The triggers for changes in community compositions are currently unknown, but infused LPS may be indirectly involved via changes in the physiology of animals in response to LPS infusion. In the present study, feed intake was not significantly affected by the LPS treatment. Thus, the alteration in ruminal pH seemed responsible for the changes. Given that changes in ruminal pH are partly responsible for altering the community structure of the ruminal microbiota( Reference Tajima, Aminov and Nagamine 53 ), it may be that Gram-negative bacterial numbers or proportions decrease in consequence of the accumulation of VFA. The present data are consistent with this hypothesis, as shown in Fig. 6. In response to the infusion of LPS, the percentage of the phylum Bacteroidetes, which is known to be the most abundant Gram-negative ruminal bacteria( Reference Nagaraja, Bartley and Fina 54 ), was linearly reduced by the LPS treatment. With regard to other physiological changes observed in the trials, the rumen temperature was not measured, but the rectal temperature increased with the rise in LPS dosage. Increased body temperature may also have an effect on the heat tolerance of respective species and thus contribute to the change in the ruminal bacterial composition.
As mentioned previously, the present study showed that the changes in microbiota composition in response to LPS infusion are accompanied by a significant increase in the concentration of SCFA in the rumen, coupled with the change in the ratio of acetate:propionate in the rumen. Because the ratio of acetate:propionate reflects the ruminal fermentation pattern, the change in the ratio of acetate:propionate in the LPS infusion group indicated that ruminal fermentation characteristics changed, which may not only be due to the changes in bacterial microbiota composition but also to the alteration of actively existing micro-organisms in the community. Examples are decreases in the proportions of Prevotella, unclassified Bacteroidales and unclassified Rikenellaceae population groups and increases in the percentages of the unclassified Clostridiales, unclassified Lachnospiraceae and unclassified Ruminococcaceae population groups (Fig. 7). Consistent with a previous study by Stevenson & Weimer( Reference Stevenson and Weimer 52 ), the present study showed that Prevotella, unclassified Bacteroidales and unclassified Rikenellaceae population groups were the predominant bacterial genera in the ruminal samples. Changes in the relative abundance of these genera may have broad physiological consequences, such as the decrease in feed degradability and propionate production( Reference Kim, Morrison and Yu 55 ). In the present study, the decrease in the abundance of the Prevotella genus in LPS-treated group implied that a possible decrease in diet degradability may occur, which corresponded with a significant decrease in the OM digestibility of the soyabean meal and alfalfa. However, the exact reason for the decrease in the proportion of the Prevotella genus after LPS infusion is not clear. However, a possible reason may be related to the lower ruminal pH that resulted from the infusion of LPS; Prevotella spp. belongs to the population of Gram-negative bacteria, and this bacterial population is known to be sensitive to environmental pH.
As functionally unknown organisms, unclassified Clostridiales, unclassified Lachnospiraceae and unclassified Ruminococcaceae were the three large groups of unclassified bacteria in the rumen. The meta-analysis has revealed that these three unclassified bacterial groups were probably competitive in the rumen and that some of their species might have an important role in ruminal feed digestion( Reference Kim, Morrison and Yu 55 ). In the present study, the increase in the abundance of unclassified Lachnospiraceae and unclassified Clostridiales in LPS treatment might relate to significant changes in the ruminal environment. It is known that significant changes in the ruminal environment such as ruminal pH are associated with alterations in the structure of the ruminal bacterial population( Reference Tajima, Aminov and Nagamine 53 ). However, with the exception of a few isolates orphaned from other genera or species, all three groups were primarily represented by uncultured bacteria( Reference Larue, Yu and Parisi 56 , Reference Brulc, Antonopoulos and Miller 57 ). Therefore, the isolation and characterisation of representative strains belonging to these groups will greatly assist in better understanding their ecology, physiology and contributions to rumen function.
Although the present study revealed that the low ruminal pH during LPS challenge was a great contributor to the changes in ruminal microbiota, other mechanisms may also be involved in the alteration of ruminal microbiota during LPS challenge. We now know that LPS acts as a stress resource by over-stimulating Toll-like receptor innate immune signalling that induces pathogenic inflammatory responses. Toll-like receptor signalling through MyD88 promotes the nuclear translocation of NF-κB and the transcription of pro-inflammatory cytokines, such as TNF and other pro-inflammatory cytokines( Reference Mani, Weber and Baumgard 58 ). As described in a review by Turnbull & Rivier( Reference Turnbull and Rivier 59 ), there is now overwhelming evidence that several cytokine families, especially IL-1, increase the levels of stress hormones, such as cortisol, in the blood. Cortisol may release into the rumen through the salvia and directly promote the growth of non-pathogenic isolates of E. coli as well as the pathogenic E. coli strain 0157:H7( Reference Freestone and Lyte 60 , Reference Freestone, Williams and Haigh 61 ). In the present study, the hormone levels in the ruminal fluid or in the blood were not detected. However, Waggoner et al. ( Reference Waggoner, Loest and Turner 11 ) showed that LPS challenge increased the serum cortisol levels in cattle. Yates et al. ( Reference Yates, Ross and Hallford 62 ) demonstrated significant positive correlations between serum and salivary cortisol values in ewes. Hence, a greater volume of cortisol may be projected into the ruminal fluid through the saliva during LPS challenge, which may be partly contributed to the changes in ruminal microbiota after LPS infusion.
Conclusion
In general, although a relatively small number of animals were used in the present study, we profiled the changes in the composition of ruminal microflora in dairy cattle after intravenous LPS challenge and observed significant changes in ruminal bacterial groups after LPS infusion. The relative populations of the phylum Bacteroidetes and the genus Prevotella decreased, while that of the phylum Firmicutes increased in response to LPS infusion. The population changes of these specific groups may be partly due to an indirect result of LPS via changing the ruminal pH value. Moreover, the present findings confirmed that LPS infusion in the circulating blood system alters ruminal fermentation and leads to a general decrease in fermentative activity. These results imply that translocated LPS during ruminal acidosis may aggravate the dysfunction of the rumen, which results in a double detriment to rumen health in dairy cattle.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S000711451400066X
Acknowledgements
The present study was supported by the National Key Basic Research Program (2011CB100801).
L. J., R. Z. and Y. L. carried out the majority of the animal studies including animal care, RNA isolation and real-time PCR. S. M. carried out the microbial data processing, analysis and interpretation. S. M. and W. Z. were responsible for the conception of the project and the oversight of the experiment. L. J. and S. M. were responsible for the writing of the manuscript.
There are no conflicts of interest to declare.













