Introduction
Listeria monocytogenes is a food-borne pathogen linked to serious illness. This disease is mostly transmitted via oral route, affecting vulnerable groups of the population such as the elderly, immunocompromised individuals, pregnant women and newborns. The high case-fatality rate of about 20% makes this pathogen a leading cause of food-borne-related mortality in humans [Reference Nyarko and Donnelly1]. Listeriosis is usually transmitted by the consumption of foods contaminated with L. monocytogenes, more frequently ready-to-eat (RTE) food such as smoked fish, cold meats or soft cheeses. RTE foods, especially those with long shelf life that support bacterial growth, are often the source of listeria infections. Such foods are usually stored in refrigerators and consumed without additional heating or cooking. In 2015, EU member states reported 2206 confirmed human cases of listeriosis [2]. In 2016, the EU notification rate was 0.47 cases per 100 000 population, whereas the rate in the Czech Republic (CR) was slightly lower at 0.45 cases. However, the number of cases is rising in both the EU and CR [3]. Nosocomial transmission of this infection is rare, but it is reported sporadically [Reference Jágrová4].
Listeriosis often occurs as sporadic cases, and small outbreaks may remain unrecognised. Molecular techniques have become increasingly integrated into routine typing practice and a molecular epidemiology approach has been confirmed as an efficient tool to investigate listeriosis outbreaks. Since 2006, four outbreaks of listeriosis have been detected in the CR. One large outbreak affecting more than 85 cases in 46 different locations, was registered between autumn 2006 and winter 2007 and was associated with the consumption of ripened cheese [Reference Vit5]. One local outbreak of listeriosis in the South Bohemian Region was reported between July 2008 and February 2009. Nine people were infected after consuming vacuum packed pork ham, which was served as a dinner in two hospitals. Four of these patients subsequently died (unpublished data). In 2009–2010, an international listeriosis outbreak was associated with the consumption of ripened sour milk curd cheese ‘Quargel’ in Austria (25 cases), Germany (eight cases) and also in the CR (one case) [Reference Fretz6].
In 2016, an increase in the number of listeriosis cases was observed in the Moravian-Silesian Region of the CR (14 cases). The incidence for this location increased to 1.15 cases per 100 000 population (CR – 0.45 cases/100 000 population). The objective of this study was the confirmation of the relationship between human cases of listeriosis and isolates from food obtained within the wide time period of 2012–2016 determined by pulsed-field gel electrophoresis (PFGE) method as a suspected epidemic cluster by whole-genome sequencing (WGS) technique.
Methods
Strains
Human L. monocytogenes isolates were obtained continuously from clinical laboratories in the whole CR. From the year 2012 to 2016, 26 human isolates belonging to indistinguishable AscI pulsotype 798/810, ApaI pulsotype 12 were found and 25 of them were sent for further typing by WGS.
Food isolates of L. monocytogenes obtained from official controls of the State Veterinary Administration (SVA) are routinely stored in deep freezers and they are processed by typing methods only during the outbreak investigation. In 2016 strains from the same period (2012–2016) have been typed. The outbreak pulsotype was detected in 14 isolates: one isolate from raw food (chicken meat), 11 isolates from RTE meat products and two isolates from the environment of the food-processing plant (Table 1). Out of them, three randomly selected isolates from RTE meat products and one isolate from the environment of food-processing plant obtained during the surveillance of outbreak in 2016 were sent for typing by WGS.
Table 1. Description of the outbreak strain isolated from food and the food-processing plant in the Moravian-Silesian Region, CR, 2012–2016 (n = 14)

Serotyping
L. monocytogenes isolates were serotyped by slide agglutination method (Denka Seiken, Japan) in combination with multiplex PCR [Reference Doumith7].
PCR clonogrouping
The clonal complex was detected with multiplex PCR [Reference Chenal–Francisque8].
Macrorestriction analysis
Macrorestriction analysis using endonuclease AscI and ApaI (New England BioLabs, USA) was performed according to the protocol Roussel et al. [Reference Roussel9] and results were evaluated by the software BioNumerics version 5.1 for analysis (AppliedMaths, Belgium).
Whole-genome sequencing
Altogether 29 strains were sent for WGS analysis in the framework of the ELiTE (European Listeria Typing Exercise) project organised by ECDC (25 strains of human origin) and also on the basis of the cooperation with AGES (Austrian Agency for Health and Food Safety) in Vienna (four strains) of food origin and environment from the processing plant producing the contaminated RTE meat products. Genomic DNA was isolated and libraries for genome sequencing were prepared according to the standardised procedure in Source BioScience. Libraries were prepared using the TruSeq DNA Library Preparation Kit (Illumina) and sequenced using 2 × 150 bp paired-end reads on the NextSeq instrument (Illumina).
Genome assembly and core genome multilocus sequence typing (cgMLST)
Genome assembly, cgMLST and data analysis using the Ridom SeqSphere+ software (version 3.5.0; Ridom GmbH, Münster, Germany) were performed at the Veterinary Research Institute (CR). The resulting FASTQ files were first quality trimmed and then de novo assembled using the Velvet assembler integrated into Ridom SeqSphere+ software. Analysis parameters: reads were trimmed at their 5′ and 3′ ends until an average base quality of 30 was reached in a window of 20 bases, and the assembly was performed with Velvet version 1.1.04 using optimised k-mer size and coverage cut-off values based on the average length of contigs with >1000 bp. Isolates were compared in seven MLST and 1701 cgMLST targets [Reference Ruppitsch10].
Results
All human isolates (26) belonged to L. monocytogenes serotype 1/2a, AscI pulsotype 798/810, ApaI pulsotype 12. On the basis of WGS results, 25 strains sent to ECDC were assigned to clonal complex CC8, sequence type ST8 and cluster type CT3239. This outbreak was reported to the European Epidemic Intelligence Information System (EPIS FWD) platform (Urgent Inquiry 378 on 5 October 2016). None of the other countries reported cases with this macrorestriction profile or WGS cluster type suggesting that this was a national continuous common source outbreak.
Human strains of epidemic pulsotype were detected occasionally in the Moravian-Silesian Region in previous years. The results of this study show that the first four human cases were recorded in 2012, six in 2013, four in 2014, three in 2015 and nine in 2016 (Fig. 1). A total of 26 cases were recorded, three of them were fatal. In the CR, mother and newborn are reported as one case. The age category 0–4 years includes two cases in twins. The infection occurred more frequently in women (non-pregnant), namely in the age category of 65 years and over (Fig. 2). Most strains of L. monocytogenes from non-maternal/neonatal cases were isolated from blood cultures (19), four strains from the cerebrospinal fluid and one strain from non-specified material.

Fig. 1. Number of outbreak cases of listeriosis, CR, in individual quarters from 2012 to 2016 (n = 26).

Fig. 2. Distribution of outbreak cases of listeriosis by age and gender, CR, 2012–2016 (n = 26).
The outbreak had a local character with the highest number of cases in the Moravian-Silesian Region (23 cases). The maximum cumulative incidence of the disease caused by this clone was recorded between 2012 and 2016 in the city of Frýdek Místek (4.22 cases/100 000 population). Two cases were also detected in the Olomouc Region (2012) and one in the South Bohemian Region (2013) (Fig. 3). Unfortunately, the patient from one geographically isolated case died and it was not possible to find a food consumption and travel history to Moravian-Silesian Region. The proportion of outbreak cases from the overall listeriosis cases in the Moravian-Silesian region was 52.3% (23/44) during 2012–2016. In Olomouc Region (2/13) and South Bohemian Region (1/12), only a small proportion of outbreak cases from overall listeriosis cases in these regions was found in this period.

Fig. 3. Distribution of human listeriosis with the outbreak strain (AscI pulsotype 798/810, ApaI pulsotype 12) at the region level, CR, in 2012–2016 (n = 26) and distribution of the outbreak strain isolated from a food-processing plant (n = 14). Dark grey circle – human, light grey circle – food-processing plant.
Epidemiologic data based on the questionnaire analysis (personal data, medical details, food habits and shopping history) did not provide the information needed to determine the vehiculum of infection for this outbreak. Epidemiological data and field investigations were conducted in parallel with the molecular analysis of L. monocytogenes isolated from food in 2012–2016. Because the results of laboratory analysis were available earlier and showed a possible link with the food-processing plant at the site under review, epidemiologists focused on the investigation at the food-processing plant, where swabs from the environment and samples of meat products were taken. Since positive food samples and L. monocytogenes of identical pulsotypes to human outbreak strains were detected, the epidemiological analysis was not continued.
Since 2012, an identical clone (AscI pulsotype 798/810, ApaI pulsotype 12) has been detected in 12 strains of L. monocytogenes by macro-restriction analysis in RTE food and raw food originating from the official controls in the Moravian-Silesian Region. The strains were isolated from poultry meat (chicken sample – raw product) and sliced RTE meat products from the same food-processing plant in the Frýdek Místek Region (Table 1). Two strains were isolated from factory-packed sliced meat products purchased in the retail market. These products had a shelf life of 2 weeks. Quantitation revealed that these samples contained 3.6 × 104 and 8.6 × 104 colony-forming units (CFU)/g of L. monocytogenes at the beginning of shelf life.
In 2016, the investigation was initiated in collaboration with local epidemiologists, which resulted in the collection of samples from the suspected food-processing plant and their examination by Supervisory Authorities. The isolates of L. monocytogenes recovered from food (seven isolates of factory-packed sliced meat products) and the environment of the food-processing plant (two) were sent to our laboratory for typing in September 2016. Isolates derived from a smoked turkey delicacy, turkey delicacy in jelly, turkey breast ham and environmental swabs showed indistinguishable PFGE profile and cluster type with human strains and strains from food isolated in previous years. A comparison of the outbreak clone with the PFGE profile detected in raw turkey meat is shown in Figure 4. By the cgMLST method, no difference was detected in most of the sequenced strains (17/29) from humans, foods and swabs from the environment of the food-processing plant under investigation. Only five strains (5/29) had three to seven different alleles (Fig. 5). Differences by region or time of isolation were not detected. In case of two strains isolated from patients from Olomouc region and one case from South Bohemian Region, no different allele was detected. No outbreak case was recorded after an intervention of control authorities at the investigated food-processing plant (Fig. 2).

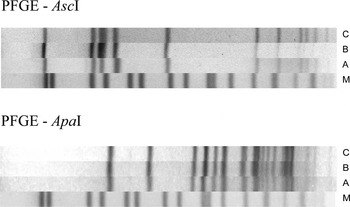
Fig. 4. AscI and ApaI pulsotypes of the Listeria monocytogenes outbreak strains, CR, 2012–2016. (A) L. monocytogenes isolate from a human; (B) L. monocytogenes isolate from a retailed RTE turkey meat product; (C) L. monocytogenes isolate from a food-processing plant (swab); (M) Salmonella Braenderup H9812 size control; PFGE – pulsed-field gel electrophoresis.

Fig. 5. Minimum Spanning Tree based on NGS allelic profiles of Listeria monocytogenes strains with AscI pulsotype 798/810 and ApaI pulsotype 12 (outbreak strain), CR, 2012–2016 (n = 29). Each circle represents an allelic profile based on sequence analysis of 1701 target genes. The numbers on the connecting lines illustrate the numbers of target genes with differing alleles. Dark grey circle – human, light grey circle (LV0815) – environment of the food-processing plant, grey circle (LV0788, LV0786, LV0671) – food.
Discussion
The first case of the outbreak strain was detected in January 2012 and the confirmation of the vehicle did not occur until the summer of 2016. The investigation of a listeriosis outbreak is difficult due to a long incubation period and a lot of possible food vehicles as a source of infection including a broad range of RTE foods [Reference Magalhaes11]. The incubation period of listeriosis is determined up to 70 days [Reference Goulet12]. Moreover, if there is a prolonged period among cases it may mean that the vehicle of infection is an RTE product distributed in the market, which comes from a single food-processing plant where persistent strain occurs.
In all human L. monocytogenes strains routine, real-time macrorestriction analysis is performed in the CR. If at least three cases of listeriosis with an identical pulsotype are recorded in one geographical region, local epidemiologists are informed of the suspect ongoing outbreak. Public health could benefit from the real-time characterisation of L. monocytogenes isolated from food. The earlier decision of control authorities to send food strains for typing could speed up the detection of unsatisfactory food products and stop this outbreak before its spread. Altogether, 26 patients mainly from the Moravian-Silesian Region were affected. The local character of this outbreak in combination with the use of molecular biology typing methods facilitated the confirmation of the vehicle of infection.
The ability of patients to recall food consumption can be problematic due to their age and the disease status [Reference Ruppitsch10, Reference Magalhaes11]. In this case, the source of the outbreak was identified on the basis of retrospective molecular typing of food isolates acquired during the official control inspection activities of the SVA. Many listeriosis outbreaks have been linked to meat product consumption [Reference Hächler13–Reference Kvistholm Jensen15]. Some authors demonstrated that strains of ST8 belonging to clonal complex CC8 are associated with meat products [Reference Henri16, Reference Fagerlund17]. Since 2013 most cases of listeriosis have been caused by strains of serotype 1/2a (58%) from clonal complex CC8 (28%) in the Czech population [Reference Gelbíčová18]. Similarly, the clonal complex CC8 has been the most frequently detected listeria CC-type in Denmark [Reference Jensen19] and Canada [Reference Knabel20]. The described outbreak was also caused by L. monocytogenes CC8 strains originating from RTE turkey meat products. All RTE food isolates tested in our study originated from sliced meat products which were packaged by the same producer between 2012 and 2016. This fact confirms that the described L. monocytogenes outbreak strain probably persisted in the environment of the aforementioned food-processing plant over several years. Persistence of L. monocytogenes in food-processing plants is one of the main root causes of human listeriosis outbreaks [Reference Ferreira21].
Foods which do not exceed the limit of 1 × 102 CFU/g for L. monocytogenes pose only a negligible risk of listeriosis development in a population of healthy humans [Reference McLauchlin22]. The Commission Regulation No. 2073/2005 on microbiological criteria for foodstuffs is also based on this limit [23]. This limit was exceeded in the case of turkey meat products confirmed as a source of the outbreak. In two meat products from the retail market, the numbers of L. monocytogenes were estimated to be of 104 CFU/g within the shelf life period. The high bacterial counts in RTE food products in the shelf life may indicate that the high-level contamination probably occurred already in the food-processing plant. In the EU, absence of L. monocytogenes is required in RTE foods able to support the growth of L. monocytogenes before the food has left the immediate control of the food business operator. This criterion applies to products before they have left the immediate control of the producing food business operator, when he is not able to demonstrate, to the satisfaction of the competent authority, that the product will not exceed the limit of 100 CFU/g throughout the shelf-life [23]. On the other hand, the US Department of Agriculture has a ‘zero tolerance’ criteria for L. monocytogenes in RTE meats overall. Maybe more strict application of EU criteria could be successful to prevent the outbreak of listeriosis caused by RTE meat products.
Macrorestriction analysis is a suitable tool for routine typing of L. monocytogenes, which enables the detection of a relationship between isolates. Nowadays, WGS is considered to be superior and highly discriminatory when allocating cases to the outbreak. The use of WGS is desirable as a routine procedure for all listeria isolates collected for typing [Reference Ruppitsch10]. The results of the PFGE method may be ambiguous in some outbreak cases. Given that the epidemiological analysis of the food consumption questionnaires did not provide the desired results, the WGS method was also used to confirm the identity of human, food and environmental strains of L. monocytogenes. For that reason, though PFGE patterns were indistinguishable in this study, WGS played an important role in the confirmation of the outbreak.
In the USA the implementation of nationwide real-time WGS on all L. monocytogenes isolates from patients, food, and the environment increased the number of confirmed outbreaks [Reference Jackson24]. In Europe, the using of cgMLST analysis is often recommended. The cut-off of ⩽10 alleles (of 1701 alleles) or cut-off of ⩽7 alleles (of 1748 alleles) has been reported that could distinguish outbreak-related isolates by the cgMLST schemes of respectively Ruppitsch et al. and Moura et al. [Reference Ruppitsch10, Reference Moura25]. All strains tested by cgMLST in our study fulfilled this limit. In fact, 59% of all sequenced strains from humans, foods and swabs from the environment had no difference, 10% of strains had one different allele, 14% two different alleles and 17% of strains had more than two different alleles (Fig. 5). On the other hand, the experience of the USA study suggests that a single cut off cannot consistently predict whether isolates will be epidemiologically related [Reference Jackson24]. Indeed, for a listeriosis outbreak linked to a single, relatively small producer, even if over several years, it may be expected to find a small number of allele differences. However, an outbreak linked to a larger producer with more food production surfaces may show greater diversity (e.g. >10 alleles) despite being linked to a single facility.
The routine use of macrorestriction analysis by PFGE of all human isolates allowed us to detect an increased prevalence of strains of the same pulsotype over the course of several years in the Moravian-Silesian Region of the CR. Retrospective analysis of L. monocytogenes isolates from food revealed the source of the infection. In this study, results of WGS and macrorestriction analysis both confirmed that all strains of human and food origin belonged to one outbreak.
Acknowledgements
We would like to thank Irena Martínková (Regional Hygiene Station of Moravian-Silesian Region, Ostrava) for her collaboration during the epidemiological investigation.
Financial support
This study was supported by the project of Ministry of Health (Prague, Czech Republic) AZV 16-31488A.
Conflict of interest
The authors declare that they have no conflict of interest.