To the Editor—Annually, more than 10,000 patients in the United States acquire an infection caused by invasive group A Streptococcus (GAS). The fatality rate of this illness is 11.7%, and many infections are transmitted person to person.Reference Flores, Luna, Runge, Shelburne and Baker 1 , Reference Nelson, Pondo and Toews 2 Outbreak investigations of postsurgical group A Streptococcus (GAS) infections can substantially disrupt surgical throughput if staff require furloughing, and they can be extremely labor intensive when surgeons practice at multiple facilities. 3 One benefit that has received little attention is the labor-saving potential that whole-genome sequencing (WGS) offers infection preventionists (IPs) when the turnaround time is sufficiently rapid to inform investigations and mitigation efforts.Reference McGann, Bunin and Snesrud 4 Here, we highlight an outbreak involving 22 surgical staff, several of whom practice at multiple facilities that often care for the same patients within a regional care network.
On day 0, patient A underwent a procedure at community hospital X, performed by surgeon I who also practices at referral hospital Y (Table 1). On day 5, patient A developed an invasive GAS surgical wound infection while at hospital X. On day 7, patient B underwent a procedure performed by surgeon II at hospital X. On day 8, patient B developed a complication requiring escalation of care to hospital Y for follow-up surgery, again performed by surgeon II. On day 13, GAS was isolated from the surgical wound of patient B while at hospital Y. The 2 GAS isolates were sent for WGS, using methods described previously.Reference McGann, Bunin and Snesrud 4 , Reference Jacob, Spilker, LiPuma, Dawid and Watson 5 Simultaneously, IP staff initiated a retrospective review of all laboratory results beginning 6 months prior to the first surgery. Involved surgical staff at all facilities were contacted to have their throats and groins swabbed. Mitigation planning was begun in case staff furloughing would be required pending decolonization.
TABLE 1 Potential Impact of an Outbreak Investigation for Surgical Site Infection due to Group A Streptococcus
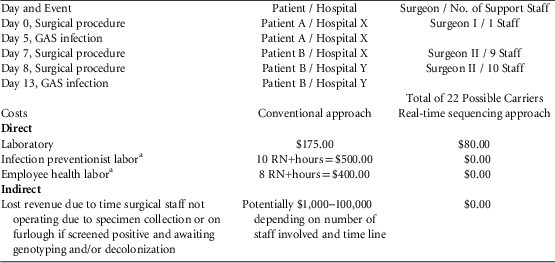
a RN+, registered nurse at 90% effort with Advanced Practitioner or Physician Oversight at 10% effort.
The core genome sequences of the 2 isolates differed by ~40,000 nucleotide changes, indicating that they were genetically unrelated.Reference Jacob, Spilker, LiPuma, Dawid and Watson 5
The WGS results were available within a week, before all staff had been swabbed and before any culture results of those that had been swabbed were available. On other occasions, results have been available in <50 hours.Reference McGann, Bunin and Snesrud 4 For this event, WGS permitted earlier termination of the investigation and faster resumption to full surgical capacity, saving time, labor, and money (Table 1). The costs in Table 1 were calculated based on material and labor costs in this regionReference Landers, Davis, Crist and Malik 6 for screening all involved operating room staff (n=22). If WGS had determined that the isolates were related, the cost would have been $80.00 more for the WGS approach compared to the conventional approach (not using WGS). When WGS revealed that the isolates were unrelated, the cost savings were substantial because surgical throughput was not slowed or disrupted, and IPs were able to devote their time and efforts to other issues. Currently, WGS has become faster, less expensive, and more informative than pulsed-field gel electrophoresis (PFGE). Furthermore, PFGE has been suggested to lead to erroneous conclusions regarding genetic relatedness among strains.Reference Salipante, SenGupta, Cummings, Land, Hoogestraat and Cookson 7 However, such timely feedback is not yet available to most hospitals; thus, IPs, surgical facilities, and patients would benefit from wider access to real-time, genome-based support.
ACKNOWLEDGMENTS
The views expressed in this article are those of the authors and do not reflect the official policies of the Department of the Army, Navy, or Air Force, the Department of Defense, or the US government.
Financial support: No financial support was provided relevant to this article.
Potential conflicts of interest: All authors report no conflicts of interest relevant to this article.





