Introduction
As the most frequent cause of the haemolytic uremic syndrome (HUS), Shiga toxin-producing Escherichia coli (STEC) (also called verocytotoxin-producing E. coli – VTEC) are the most feared diarrhoeagenic E. coli. HUS is characterised by the sudden occurrence of microangiopathic haemolytic anaemia, thrombocytopenia and renal insufficiency. Most cases are preceded by a prodromal episode of diarrhoea caused by the STEC infection, missing in the so-called atypical HUS cases, due to complement dysfunction [Reference Piérard1]. In Belgium it is mandatory to report STEC infections to the regional health inspection authorities. These authorities investigate each notified case separately and decide which measures have to be taken to prevent further spread of the infection and perform outbreak screening and investigation if necessary. The criteria used for STEC case definition are those provided by the European Centre for Disease Control (ECDC). All Belgian clinical laboratories can send stool samples, rectal swabs from HUS patients, faecal cultures on agar and strains suspicious for STEC to the National Reference Centre (NRC) for diagnosis and strain typing free of charge. Referral of specimens to the NRC is voluntarily, but is highly recommended [Reference Muyldermans2].
It can be difficult for the health inspection authorities to determine whether a case of STEC infection has to be excluded from a group (e.g. children's day care centres) or an occupation (e.g. food handlers) and whether it is necessary to screen for asymptomatic carriers. To facilitate the decision making process regarding the management of STEC infections, information about the risk factors for the development of HUS can be helpful. Previous studies identified the presence of the STEC virulence genes stx2 in general, subtypes stx2a and stx2d more specifically, and eae, as well as young and older ages of the patient as risk determinants for HUS development. Since the large-scale STEC O104:H4 outbreak in 2011, the rare combination of enteroaggregative virulence genes and stx2 is also considered as high risk [Reference Scheutz3–Reference Scheutz6].
In order to provide insight in the risk determinants for typical HUS development in Belgium, we performed statistical multivariate analyses on a dataset of STEC strains isolated at the Belgian NRC STEC between 2011 and 2016. Based on the results, we propose an adapted STEC typing algorithm and risk classification of STEC strains to provide our local public health authorities with the most useful information.
Materials and methods
Data collection
Laboratories sending samples to the NRC STEC for diagnosis and strain typing are asked to fill out a request form. This form contains questions about the sample (type of sample, sample date) and the patient (age, sex, clinical manifestations, date of onset, recent travel, suspected vehicle, cluster or sporadic case). At the NRC, attempts are made to isolate a STEC strain from each stx-positive sample by screening up to 20 single colonies for the presence of stx genes. Typing of the stx-positive strains is performed in different phases. Biochemical characterisation, motility testing, detection of the top six O-serogroups (O26, O103, O111, O121, O145, O157); antibiotic resistance testing; and PCR for stx1, stx2, eae, hlyA, aaiC and aggR virulence genes are performed immediately as described previously [Reference Buvens7, Reference De Rauw8]. In order to quickly identify outbreaks of STEC O157, the most common O-serogroup associated with outbreaks in Belgium, IS629-typing of STEC O157 is also performed as soon as possible [Reference Ooka9]. Additional serotyping using sequence-based typing of the gnd-gene locus and stx-subtyping is performed in batch every 3 months [Reference Gilmour10, Reference Scheutz11]. Suspicion of outbreaks by other serotypes is confirmed by pulsed field gel electrophoresis or whole genome sequencing analysis. The typing results of each strain, as well as the according sample and patient information, are collected in an anonymised database. Only one strain per patient per infection episode is stored.
Strains selection
Only STEC from Belgian patients with known HUS status (HUS or non-HUS) isolated between 2011 and 2016 were included. In case of known clusters of infection or outbreaks, only one strain per outbreak, derived from the patient with the worst outcome, was selected. Using these criteria, 411 strains were selected for statistical analysis.
Statistical analyses
Statistical analyses were performed using the IBM SPSS Statistics software 24 and Microsoft Excel. Univariate logistic regression analyses were performed to assess the risk for HUS development of different patient and STEC strain characteristics. Variables with a P value < 0.05 were considered statistically significant. Positive predictive values (PPV) and negative predictive values (NPV) were computed for statistical significant variables in relation to the presence of HUS (Supplementary Tables S1–S3). Three different multivariate logistic regression models (A, B, C) were used on variables selected on the basis of their relevance and significance in univariate analyses. In model A, a distinction was made between stx1 (with or without stx2) and stx2 (with or without stx1) positives. In model B, stx was categorised in stx1 (not stx2) positives, stx1 + stx2 positives and stx2 (not stx1) positives. In model C, significant stx1 and stx2 subtypes were taken into account.
Results
Univariate analyses
Univariate logistic regression analyses showed the following characteristics to be statistically significantly correlated to the development of HUS: patient age categories ⩽5, 6–12 and ⩾75; STEC serotypes O157 and O145; and STEC genes stx2, subtype stx2a more specifically and eae. Presence of the stx2a gene had the best PPV and NPV, 38.7% and 95.4%, respectively. The following variables were significantly correlated to a reduced risk for HUS development: STEC fermentation of sorbitol and production of β glucuronidase; and presence of the stx1 gene (Table 1, Supplementary Tables S1–S3).
Table 1. Statistical significant results of the univariate logistic regression analyses

a The total number of patients with known age was 409; the total number of strains was 411 for all characteristics except for β glucuronidase production which was known for only 403 strains.
b CI, confidence interval.
c PPV, positive predictive value.
d NPV, negative predictive value.
Multivariate analyses
Analyses of the significant and relevant variables in multivariate logistic regression models excluded the STEC O serogroups O26, O145 and O157; sorbitol fermentation; and β glucuronidase production as significant risk predictors. Age categories ⩽5, 6–12 and ⩾75, and the stx2 gene remained significant risk determinants in all three models (Table 2, Supplementary Tables S4–S6). Presence of the eae gene was significantly correlated to a higher risk in two out of the three models. Detection of the stx2a gene had the highest risk for HUS development (OR 29.6, 95% CI 7.0–125.1) (Table 2, Supplementary Table S6). Presence of the stx1 gene, regardless of the subtype, had a lower risk for HUS development. Presence of the stx1 gene without stx2 appeared to encompass a lower risk than the combined presence of stx1 and stx2, while the presence of stx2 had a higher risk than both genes (Table 2, Supplementary Tables S3 and S5).
Table 2. Statistically significant results of the multivariate logistic regression models A (stx1 (with or without stx2) vs. stx2 (with or without stx1) positives), B (stx1 (not stx2) positives, stx1 + stx2 positives, vs. stx2 (not stx1) positives) and C (significant stx1 and stx2 subtypes)
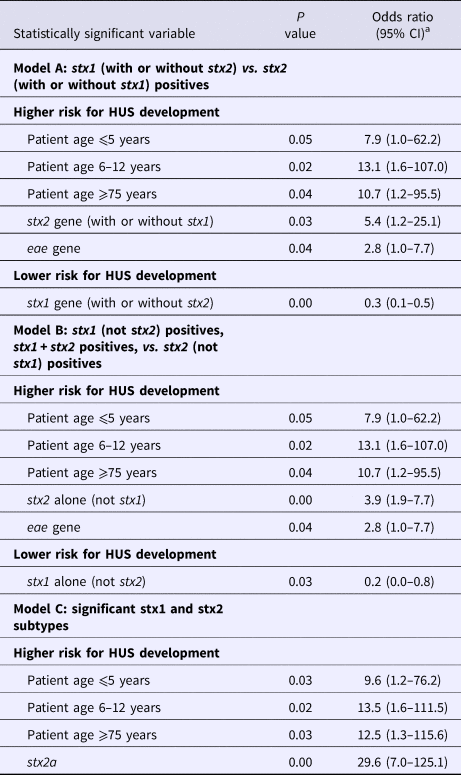
a CI, confidence interval.
Discussion
Univariate logistic regression analyses showed patient ages ⩽12 and ⩾75 years, STEC serotypes O157 and O145, the Shiga toxin stx2 gene in general and the stx2a subtype more specifically, and the virulence gene eae to encompass a higher risk for HUS development. However, only patient age and the stx2 gene remained significant risk determinants in all three multivariate models. The presence of the eae gene was significant in the multivariate models A and B, but not in model C. This indicated that the presence of stx2a was the most important risk marker of the included strain characteristics. Unfortunately, we were not able to make statistically significant risk predictions about other stx2 subtypes based on our dataset.
Sorbitol fermentation and β glucuronidase production were protective markers for HUS development in the univariate analyses. This is in correlation with the fact that many non-O157 strains are sorbitol fermenters and β glucuronidase producers (178/204 and 175/198, respectively), while the majority of STEC O157 do not (203/205). After multivariate logistic regression analyses however only the stx1 gene stayed statistically significantly correlated to a lower risk for the development of HUS. Because STEC strains can possess a combination of stx1 and stx2 genes, we conducted multivariate logistic regression analysis B. This revealed that strains positive for stx2 but not for stx1 had a higher risk for HUS than those possessing a combination of stx1 and stx2 genes, indicating stx1 can tone down the effect of stx2.
The results of our study are not new; old and young age, and the genes stx2a and eae have been found to be statistical significant risk factors for HUS development in other studies before. However, previously published data often included a lower number of cases and were focussed on a specific country or region [Reference Byrne12–Reference Haugum14]. For this reason, they cannot be extrapolated to other countries or regions without further research. The more data from different countries get published, the more insight will be achieved in the possible risk factors for severe disease development associated with STEC infection. Furthermore, unlike previous studies, we have actually used the results of our study to turn around the STEC virulence typing scheme at the Belgian NRC. Upon the time of this study, all STEC isolated at the Belgian NRC were immediately characterised for the presence of the six most important STEC O serogroups (O26, O103, O111, O121, O145 and O157) and the virulence genes stx1, stx2, eae, ehxA, aaiC and aggR; while stx subtyping was only performed every 3 months. Based on the presence of the eae and ehxA genes, strains were reported as being ‘typical EHEC’ when both genes were present, while they were reported as ‘atypical EHEC’ when they lacked one or both genes. Studies showed this classification was not clearly correlated to the outcome of the infection, especially with the risk for HUS development [Reference Nataro and Kaper15]. Because the results of our statistical analysis confirmed that stx subtype is the most important indicator of HUS, we have proposed a new typing algorithm that requires an equal amount of labour as our present one. Our analysis showed stx2a to be the only stx subtype with a statistical significant higher risk for HUS development and this will be detected and reported immediately. Because stx2d is a rare subtype in Belgium (only 8/411 strains (Supplementary Table S3)) we were not able to tell something with statistical significance about this subtype. However, stx2d is considered as a high risk subtype in other studies, and for this reason, we decided to also include this subtype in our first-line typing [Reference Scheutz3]. Because the role of ehxA in HUS development was not significant in our statistics, this gene will only be detected quarterly on groups of isolates. Detection of the rare enteroaggregative genes aaiC and aggR will only be performed promptly for eae-negative strains from HUS patients or outbreaks (Fig. 1). Although O serogroups do not appear to be important HUS risk factors, we will still report them, as they are informative and easy to obtain by agglutination of colonies. Biochemical identification of the strains, including sorbitol fermentation testing, will still be performed immediately as well. These tests are easy and cheap and allow us to detect sorbitol-fermenting STEC O157, which are rare in Belgium, but have been the cause of severe disease and outbreaks in Germany and other European countries [Reference Brandal4, Reference Werber16, Reference Vygen-Bonnet17]. By implementing this new virulence typing algorithm, we will be able to provide the physicians and health inspection authorities with a risk classification for the development of HUS which is primarily based on the stx (sub)type (Table 3). We believe this classification will contribute to optimal infection control management, as for instance during the decision making regarding children's day care centres exclusion policies. Nevertheless, we are aware of the fact that only a limited number of patient and strain characteristics were studied here and that a classification based on the stx subtype only also has its limitations. A recent study in Norway identified the non-LEE effector protein nleH1-2 as an additional potential independent risk factor for HUS development [Reference Naseer18]. In the future, more extended virulence profiling could be done by using whole genome sequencing of STEC. We also acknowledge the fact that there is a possible bias regarding the cases that could be included in this study. As the NRC, we are dependent on the external laboratories and practitioners who refer their samples to provide us with the clinical information of the patient. Unfortunately, we do not always receive information about the disease of a patient diagnosed with STEC, and these strains could not be included in this study. Finally, it should be noted that every STEC regardless of its profile has the potential to cause severe disease as several host factors including the patient's genetic background already have shown to play a role in the susceptibility to and severity of the disease [Reference Taranta19].
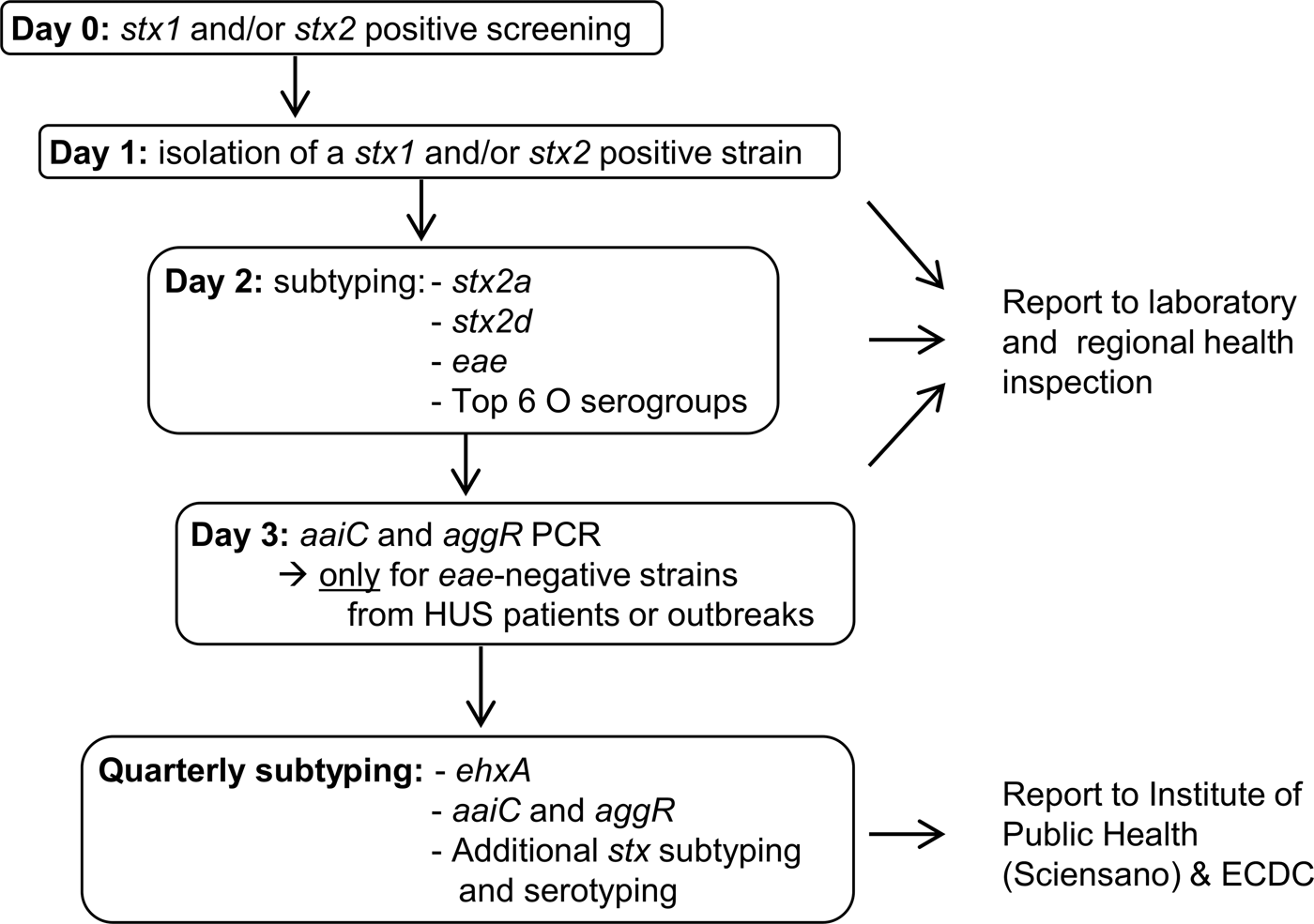
Fig. 1. Proposal of a new STEC virulence typing algorithm at the Belgian NRC STEC.
Table 3. Proposal of a new risk classification of STEC strains for the development of HUS

The presence of eae or aaiC/aggR is also associated with a higher risk for HUS development.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268818002546.
Acknowledgements
Part of this work was performed in the frame of the Belgian National Reference Centre for STEC supported by the Belgian Ministry of Social Affairs through a fund within the Health Insurance System.
Conflict of interest
None.
Ethical standards
Epidemiological data were collected anonymously in the frame of Decision No 2119/98/EC of the European Parliament and of the Council, concerning the epidemiological surveillance and control of communicable diseases in the Community, as completed by Decision No 1082/2013/EU.








