Introduction
Modern imaging technology has enabled nanoscale imaging to become a routine process. As imaging technology improves and image size increases, application measurement needs become more demanding and complex. These needs can be illustrated in the application areas such as nanoparticle detection during a manufacturing process or dye particle characterization in printing processes. For example, nanoparticles released during manufacturing processes can impact worker safety, and the number, size, morphology, and distribution of the released particles can help in better assessing the hazards associated with these processes and minimizing the risk to workers. In printing processes, final dye particle size and distribution in the coatings layer or paper fibers can vary depending on the environmental conditions. Although we are able to image nanoscale features and phenomena with relative ease, application-specific measurements such as nanoparticle morphology or dye particle size distribution over large physical scales are non-trivial because of the complex computations and associated requirements imposed on both software and hardware. If such measurements are made possible, scientists could gain a better understanding of how nanoscale features are spatially distributed and how they interact with macro and mesoscopic scale environments.
Several advanced microscopes have the capability to automatically acquire overlapping images of small fields of view [Reference Eberle1–Reference Brantner2]. This capability provides a way to image a large field of view (FOV) of a centimeter-sized specimen and then take measurements at multiple physical length scales. However, automated specimen scanning yields thousands of small FOVs (or image tiles) that need to be pre-processed into one large FOV, visualized during explorations, and measured to extract useful information. The large FOV is typically of the order of giga-pixels. A set of large FOVs over time, across wavelengths, or with varying z-dimensions can reach terabyte (TB) storage levels. These TB-sized 3D data sets pose challenges to on-the-fly measurement, interactive exploration, and collaborative discovery. In other words, while it is possible to collect the necessary images at the requisite resolution, it is hard to examine these images as a whole and interpret the data collaboratively to understand the physical phenomena under study.
This article introduces a web-based system that enables a user to explore and measure objects of interest at multiple length scales interactively and collaboratively after thousands of small FOVs are pre-processed into a set of large FOV images. We illustrate the use of the system on gigapixel images of aerosolized nanoparticles and dye particles on printing paper. Other larger data sets that have been processed using the systems are available at isg.nist.gov.
Materials and Methods
Electron microscopy test data
We imaged a specimen of nanofibers air sampled onto a Si wafer using an FEI Helios NanoLab 650+ (Hillsboro, OR) focused ion beam scanning electron microscope (FIB SEM). Small FOVs of a specimen were acquired and stored as TIFF images using FEI’s MAPS correlative microscopy software. The small FOVs overlap by 10% in horizontal and vertical directions and form a grid of 40 × 59 image tiles. Each FOV image is 6.3 MB (6 MiB1, 3,072 × 2,048 pixels, 8 bits per pixel, 49 × 49 nanometer pixels). Another similar set of test images was acquired by imaging dye particles on printing paper with the same microscope. The small FOVs have an overlap of 10% in horizontal and vertical directions, and they form a grid of 20 × 20 raw image tiles of the same pixel size. In both data sets, the goal is to characterize size, shape, and spatial distribution of these small particles in a spatially large specimen.
Main methods
We designed a web-based (client-server) system to handle the following tasks: (1) uploading collections of small FOVs to a web application with access to computer cloud resources, (2) configuring the computations to assemble a large FOV, (3) viewing and exploring the large FOV via an interface for spatial pan-zoom and time/spectral-sweep, and (4) executing interactive and collaborative measurements over the large FOV to optimize, validate, and verify results of automated parameterized computations. Figure 1 illustrates user interactions with the web system and the steps in the processing assembly part of this client-server system.

Figure 1 Overview of the data and analysis workflows showing the possible interactions by multiple users. First, a user uploads images via his browser and specifies parameters for assembling collections of small FOVs into one large FOV. The assembly consists of a minimum of three steps, such as tile stitching, intensity scaling to 8 bits per pixel, and pyramid building. Next, a user configures his views of multiple channels (overlays of multiple pyramids) and multiple time points (pyramid sequences), and then takes intensity measurements via interactive display, distance measurements using a scale bar or two-point mouse clicks, and object count measurements by running filtering and connectivity analysis tools in a browser.
1. Upload user interface (UI): A user can drag and drop folders with files or individual files one by one to a browser UI to upload image collections. File types are automatically classified as image or text based on their suffix (MIME type).
2. Large FOV assembly: The assembly of a large FOV requires a sequence of computational steps running on the server (top of Figure 1). The type of input images (small FOVs) determines the choice of steps and their order. A user is able to set up parameters via one or more user interfaces in a browser and launch computational steps in any order as long as input/output data types match. The minimum number of computational steps includes (1) the estimation of a stitching vector containing the global image tile positions for all the small FOVs, (2) intensity scaling if the acquired small FOV intensities range outside of 0 to 255, and (3) multi-resolution pyramid building.
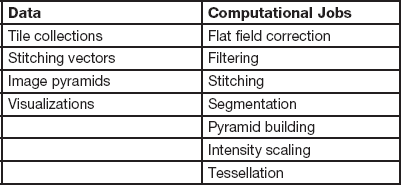
The intensity scaling is necessary because current browsers only support images with 8 bits per pixel. The image pyramid shown in Figure 2 is constructed to avoid limitations of typical network resources. The pyramid representation is useful for transmitting to end users only the portions of the image that are being viewed. In addition to enabling interactive pan-zoom viewing, the pyramid construction from small FOVs also avoids the challenges associated with constructing one large multi-giga-pixel file that might not fit to RAM. Additional computational steps can be launched as needed. All available computations are listed in Table 1. All metadata coming from microscope acquisitions and intermediate workflow computations are hyperlinked with the resulting data, as computational steps are applied to assemble a large FOV. The metadata are stored following the Open Microscopy Environment (OME) file format (OME Data Model, OME-XML, and OME-TIFF) and allow traceability of the results to data acquisition and computational steps. Figure 3 illustrates user access to either Data types or computational Jobs listed in Table 1 via the drop-down menus. The figure also shows measurement tools and an interactive pan-zoom view of one large FOV that was selected from the resulting Data types.

Figure 2 An illustration of multi-resolution pyramid representation of images of dye particles on printing paper. The vertical axis corresponds to the level of details. The red line in each horizontal plane denotes spatial partitioning into overlapping pyramid tiles.

Figure 3 The interface to “Data” and computational “Jobs” functions at the top of the web application is via drop-down menus. The accordion menu on the left lower side contains filters, connectivity analysis to obtain counts of objects, and distance measurements that are linked to the viewing area on the right side. Any change of filter parameters will be reflected in the image display while different areas at multiple resolutions are explored using pan and zoom, and distances are measured by activating the measurement tool and using mouse clicks.
Table 1 A list of data and computational jobs accessible via web user interfaces.

3. Interactive viewing and exploration of large FOV: Interactive viewing is based on transferring pyramid images from a server to a client according to a user-chosen level of detail (zoom) and spatial location (pan). Multi-resolution image pyramids have been previously used in the context of “Virtual Microscopy” [Reference Romero3] as efficient representations for transmission and viewing of large images. We enhanced the viewing experience by displaying calibration information for the intensity and zoom level at the mouse cursor location. In addition, a user can bring together all the datasets for transparency-based overlay, multi-channel viewing, and measurement extraction via visualization configuration UI.
4. Interactive measurements in a browser: Current computational resources do not allow for the processing, transfer, visualization, and interactive measurements of TB-sized images. However, scientific explorations and measurements rely on data presented on a computer screen. Our approach is to process and visualize measurements of only the data being viewed in a browser. This approach avoids transferring all data between server and client, while leveraging hierarchical partitioning of a large FOV into its pyramid representation. It also assumes that the client/browser will be able to perform computations in near-real time over screen-size images, a capability present in most web browsers. Thus, measurement operations are applied either directly to the pyramid tiles or to rendered pyramid tiles inside of a HTML canvas element. By operating directly on pyramid tiles, a user can zoom and pan while visually inspecting the updated results of measurements.
Results
We illustrate how to assemble a large FOV of aerosolized particles from 40 × 59 FIB SEM image tiles, explore the spatial distribution of particles, and measure them using the web-based solution.
1. Assembly: After uploading the aforementioned data to the web system, the collection of image tiles is processed by a set of user-specified steps (that is, a computational workflow). The workflow consists of executed Jobs in the menu, such as (1) “flat field correction jobs” using Gaussian filtering (kernel=120) followed by subtraction from the original (see Figure 4), (2) noise “filtering jobs” using the Gaussian kernel of size 3×3, (3) “segmentation jobs” by Empirical Gradient Segmentation (EGT) [Reference Chalfoun4] and background removal based on EGT segmentation, (4) “stitching jobs” using the background removed input tiles and the MIST algorithm [Reference Blattner5] derived from existing direct image alignment methods [Reference Szeliski6–Reference Kuglin and Hines8], and (5) multi-resolution “pyramid jobs” using the stitching vector and flat field corrected tiles (layer 1) and segmentation (layer 2). All workflow steps are also listed in Table 2. The steps are described in terms of their inputs, outputs, and operations, and classified based on the client or server location of computations.

Figure 4 Stitched images of small FOVs without flat field correction (top left) and after flat field correction (bottom left). Both images on the left have been contrast-enhanced via histogram equalization on the right side to show the differences. Each FOV image is 3,072 × 2,048 pixels (148.992 × 99.328 micrometers). The stitched image is 223,836 × 122,577 pixels (10.856 × 5.945 millimeters).
Table 2 Description of basic workflow computations that can be applied to input images.

The design of a workflow sequence depends on the signal-to-noise ratio (SNR) of acquired images and on the time and computational resources allocated for the measurement tasks. Figure 4 shows visual comparison of stitching with and without flat field corrected small FOVs and the size of one FOV with respect to the imaged size of a specimen. The standard deviation of image with flat field (14.5) becomes about 1.5 times smaller than the standard deviation without flat field correction (22.2). The computations can be executed on distributed resources managed by the server to meet execution time constraints. In the “worst” case of a server being a single virtual machine with 8GB of RAM (Processor: dual core Intel Xeon 2.93 GHz; Operating system: Ubuntu 12.04 and VMware Hypervisor vendor), the overall computation time was 19 h and 15 min.
2. Interactive exploration: To explore the assembled images (that is, view the pyramids), a user selects from the Data menu “Pyramids” to view one layer or “Visualizations” to configure Deep Zoom with two or more layers. Figure 5 shows an overlay of flat field corrected tiles (layer 1) and segmentation (layer 2) with the transparency defined by a slider bar under “Layer composition” (left position: layer 1, right position: layer 2). The visual exploration using a mouse takes advantage of pan and zoom (mouse click-drag=pan, scroll=zoom), pixel level information (mouse hover), and further inspection after selecting an area of interest using pan-zoom and downloading it (screen recording option in the left pane).

Figure 5 Deep Zoom visualization of an overlay of flat field corrected assembled tiles into a large FOV and its segmentation with the transparency defined by a slider bar under “Layer composition”. The FOV contains rod-like (high aspect ratio) particles of interest and round polymer fragments collected during the sampling process.
3. Measurements of particles: The interactive measurement tools in a browser are embedded in the Deep Zoom visualization in the left panel under Filtering, Connectivity Analysis, and Distance Measurements. Filtering options include typical operations for image enhancements including thresholding and edge detection. Figure 6 shows the list of available filters and the interactively updated results image, as a Sobel edge filter followed by Brightness filter are applied and the Brightness parameter is varied.

Figure 6 A list of available filters invoked by “Modify filters” button. The order of filters applied to pyramid images can be changed by dragging the filter name in the “Selected filter” pane.
Given a good separation of particles from their background by either running a Segmentation job or applying a sequence of filters under Filtering, one can invoke the Connectivity analysis by clicking on “Run analysis” button. Figure 7 shows a result for the segmented mask selected as the input and viewing area shown in Figure 5. The resulting information can be used to assess a spatial density of particles of a particular area in pixels {[red, 1352], [green, 1338], [blue,1161], [yellow, 1035], [pink, 825]} or in nm2 by multiplying the pixel area by 6534 nm2 (the pixel dimensions 48.5 nm × 48.5 nm divided by the zoom level 0.6 × 0.6 equals to 6,534 nm2). If the objective is to measure the length of particles (or any other objects of interest) then distance measurements can be obtained by mouse clicks after checking the check box “Active measurement.” Figure 8 shows distance results for the elongated nanofibers with color-coded points and their corresponding measurements in SI units {12, 10, 8, 8, 10, 6, 5, 5, 6, 7} [μm] (average =7.7 μm, standard deviation = 2.36 μm).

Figure 7 When the button “Run Analysis” is pressed, an image is binarized into black and white pixels by thresholding (that is, white pixel if above and black pixel if below intensity threshold), all neighboring pixels of the same binarized pixel are connected into regions (connectivity analysis), and a unique number/color is assigned to each connected region. The right panel presents color-coded connected regions sorted by area and total count of regions viewed in a browser window (in this case, the viewing area is shown in Figure 5 and consists of rod-like particles of interest and extraneous round polymer fragments).

Figure 8 Distance measurements are obtained by mouse clicks on end points of nanofibers. The measurements are recorded in the left pane and can be deleted or downloaded (clicked point coordinates and distance).
Discussion
A combination of assembly computations yielded a large FOV and proved to be robust to varying SNRs, feature types, and amount of overlap in each input collection of thousands of small FOVs. While we did not present numerical robustness results, we have performed extensive evaluations of the stitching method applied to large and diverse datasets (5 microscope types, 4 imaging modalities, 8 types of content, 10–70% range of image overlaps, and a wide range of grid sizes from 5 × 5 to 70 × 93). In addition, we have accelerated the stitching algorithm on hybrid CPU/GPU hardware [Reference Blattner5] and compared its performance during a stitching challenge hosted at NIST in 2015 (https://isg.nist.gov/BII_2015/webPages/pages/stitching/Stitching.html).
The web assembly computations and interactive online measurements have been tested on the NIST intranet as the system is open to all NIST scientists. We also provided a public access to several data sets for interactive analyses at https://isg.nist.gov/deepzoomweb/data/materialparticlesdistribution after the raw images were pre-processed. While accessing the data sets across the Atlantic or the Pacific, the bandwidth affected the image retrieval, but the rendering was still interactive. Once images are rendered the measurement interactivity is intact regardless of the geographical location because measurements are executed in a web browser. For parties interested in deploying and using the web system internally, we posted the deployment instructions and the code for uploading, preprocessing, and interactive measurements at https://isg.nist.gov/deepzoomweb/resources/csmet/pages/web_image_pipeline/web_image_pipeline.html.
Conclusion
We have designed a client-server system for interactive discoveries by scientists interested in multiple length scale measurements of a specimen. We illustrated the use of the web system for measuring aerosolized nanoparticles and provided access to data and code on our NIST web server. In the future, we plan to automatically select the optional pre-processing computations that affect the stitching performance and expand the set of processing operations. We are also interested in evaluating the accuracy and uncertainty of each large FOV assembly computation in order to characterize the accuracy of derived measurements from the large FOV.
Disclaimer
+Commercial products are identified in this document in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by NIST, nor is it intended to imply that the products identified are necessarily the best available for the purpose.