In 1949, geneticist and serologist William Boyd wrote to the American Journal of Human Genetics to praise recent research on the Rhesus blood groups and affirm their superlative importance for clinical medicine, anthropology and human genetics. But he warned that before making use of the vast abundance of blood-group data, researchers would need to take particular care to ‘understand the symbols in which they are expressed’. This, he regretted, was no simple task, owing to the existence of ‘two rival systems of nomenclature, neither of which has as yet won exclusive acceptance’.Footnote 1
Boyd was intervening in a dispute that had been simmering in the scientific and medical press for almost five years. The controversy's chief protagonists were American serologist and geneticist Alexander Wiener, who had first defined the Rhesus groups in 1941, and the British geneticists Ronald A. Fisher and Robert Race, who, three years later, had proposed a novel hypothesis about the genetics of the system and an entirely new nomenclature to go with it. As the Rhesus system grew in complexity over the next decade, the Wiener and Fisher–Race nomenclatures grew too. They were used and modified by doctors, clinical pathologists, immunologists, geneticists and anthropologists, who debated their merits and suggested alternatives. Meanwhile, official bodies held several international meetings to standardize the nomenclature one way or the other. Yet in some respects the dispute was never resolved, and though the controversy faded, both nomenclatures – or modified versions of them – continue to be used to this day.
Several scientists, physicians and historians have recounted the story of the Rhesus controversy.Footnote 2 Most view it first and foremost as a dispute over genetic and immunological theory, and explain its longevity by citing Wiener's tenacious defence of his own ideas. In the most thorough account, Pauline Mazumdar gives an insightful analysis of the theoretical commitments of the main protagonists, from which I draw extensively.Footnote 3 But the main stimulus for my paper is Mazumdar's brief suggestion that in the end the fates of the competing nomenclatures were probably determined less by their ‘immunogenetic implications’, and rather more by their ‘practical usefulness’.Footnote 4 The Rhesus controversy is one of the more public disputes over nomenclature in the history of science, and offers rare access to the functions of, and demands on, nomenclatures in the laboratory and clinic. This paper uses the controversy to explore the practical and epistemic qualities of nomenclatures, and to analyse the range of media and contexts in which they were expressed.
I draw on recent analyses that have framed diagrams, notations and formulae as part of the material culture of work on paper. These accounts show that work traditionally regarded as ‘theoretical’ demands practical skills, technologies and ‘paper tools’, no less than practices at the laboratory bench.Footnote 5 Just as the design of instruments shapes the outcomes of experiments, so the syntax of paper tools – their form, rules for operation and manoeuvrability – shapes the products of theoretical work.Footnote 6 I pay particular attention to the ways that nomenclatures functioned not just as representation devices, but also as instruments deployed on paper to generate and extend theory.
While nomenclatures and diagrams have been taken seriously by historians of physics and chemistry, they have attracted less attention from historians of biology. But during its first half-century at least, genetics was largely about doing work on paper. Geneticists collected data from experimental crosses or family surveys, which they assembled into tables, inscribed as symbols, and manipulated on paper according to operational rules.Footnote 7 The practices for working out these genetic rules on paper had algebraic qualities similar to the nineteenth-century chemical formulae discussed by Ursula Klein in her book on ‘paper tools’.Footnote 8 But despite the central generative role of nomenclatures in genetic protocols, we have little analysis of their functions. Exceptions include work by Robert Olby, and more recently James Griesemer, who use Gregor Mendel's notations to trace his theoretical commitments or ‘research style’.Footnote 9 Elsewhere, Raphael Falk, Sara Schwartz and Robert Kohler have written about a ‘notational crisis’ that forced Thomas Hunt Morgan to abandon neo-Mendelian schemes and develop his new project of genetic mapping.Footnote 10 These accounts presuppose a performative, generative role of inscriptions, and tangentially imply that notation might constrain conceptual change. But although we have detailed analyses of genetic maps – as representations, as devices to materialize questions, and as discursive technologies – we know relatively little about the practices of drawing genetic crosses or rendering pedigrees.Footnote 11 And while several accounts have studied nomenclatures to illuminate the conceptual issues at stake in the study of heredity, none have followed the fates of nomenclatures themselves.
The study of human heredity was particularly difficult ground for negotiating nomenclatures. Researchers studying human inheritance often focused on medically defined traits, so their interpretation was shaped by practices in medicine and other research fields. Blood groups are a consummate example of this; during the interwar period they were made into exemplary Mendelian traits for geneticists and immunologists, but were also deployed extensively in the process of transfusion therapy, in anthropological studies of race, and in forensic and paternity disputes.Footnote 12 All of these workers depended on one another for techniques, samples and data. Moreover, blood groups could not be seen and manipulated directly, rather they were defined when samples were mixed – when red cells clumped together, or agglutinated, samples were ascribed to different groups – and took on material forms primarily as inscriptions on paper. Following these inscriptions we can trace how the Rhesus groups became progressively more elaborate in the laboratory as more types emerged with increasingly subtle genetic architectures, while they led rather simpler lives in the hands of doctors and transfusion workers. In this paper I discuss how the disputed nomenclatures were articulated in different kinds of laboratory and medical environment and how users' choices corresponded to different practical activities.
This leads me to a more general contribution to the history of representation in scientific practice by extending recent studies of relations between different media.Footnote 13 Sifting through the literature of the Rhesus controversy reveals that competing nomenclatures were ascribed different virtues as they moved between material forms. Scientists and doctors discussed how blood groups and genotypes might best be written down, printed or spoken out loud.Footnote 14 I argue that to understand the dynamics of nomenclature use we might take seriously how they were articulated – in handwriting, in print and in speech.
First I outline the early relationship between the competing nomenclatures and the theoretical commitments of the main protagonists. I then turn to how the nomenclatures functioned as organizational tools in serology and genetics, emphasizing their productivity as objects for doing experiments on paper, particularly for genetic research. Moving beyond the laboratory notebook, I draw attention to how researchers and clinicians marshalled arguments about how nomenclatures should be printed, written and spoken in the laboratory and clinic. Finally I discuss how competing nomenclatures also reflected commitments to different objects in different disciplinary contexts, and end with some reflections on what these observations reveal about the protracted nature of the controversy itself.
Discovery and early history of the Rhesus groups
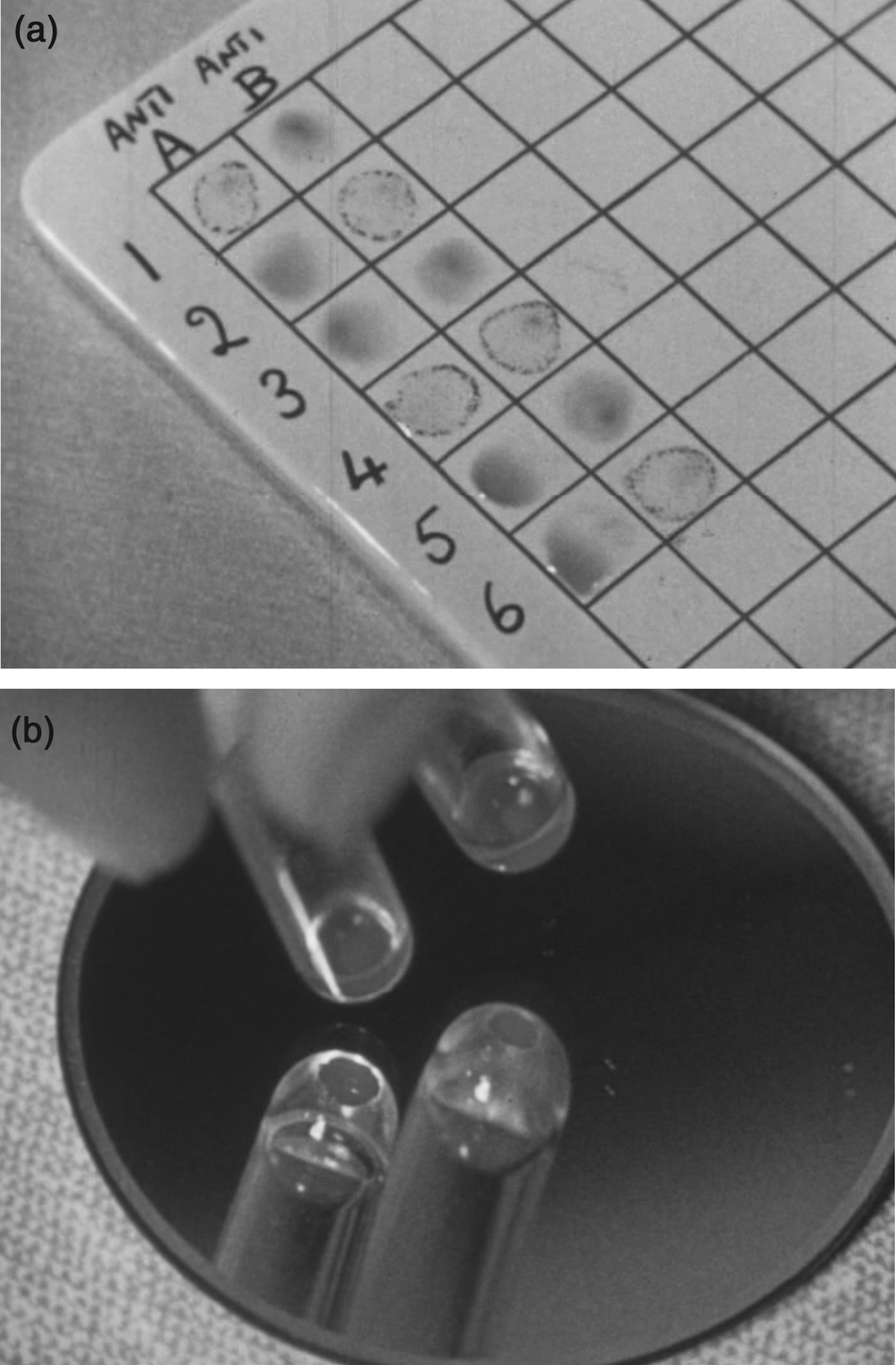
The Rhesus groups were first defined in 1940 by Karl Landsteiner and Alexander Wiener. Landsteiner – who in 1930 had won the Nobel Prize for his discovery of the ABO groups – was an immunologist at the Rockefeller Institute, while Wiener was associated with the Jewish Hospital of Brooklyn, the Department of Forensic Medicine at New York University, and the Office of the New York Chief Medical Examiner. Positioned at the nexus between the clinic, the medical research laboratory, and the genetics community, Landsteiner and Wiener had discovered the first Rhesus antiserum during a series of experiments using blood-group agglutination to determine evolutionary relationships among primates.Footnote 15 Researchers understood agglutination to be caused by antibodies in one sample binding to cell-surface antigens of the other, so in practice the group of a new blood sample was inferred by mixing it with a series of antibody-containing reagents, or ‘antisera’ (Figure 1). The researchers had found that rabbit antisera immunized with blood from Rhesus monkeys caused agglutination in 85 per cent of human blood samples, and soon after identified a human serum that caused exactly the same pattern of reactions.Footnote 16 They called the blood samples that agglutinated with these sera ‘Rh+’, and those that did not ‘Rh–’.Footnote 17

Figure 1. Figures 1a and 1b. Stills from film demonstrating ABO and Rhesus blood-grouping technique. (a) ABO tests tended to be carried out on a white ceramic slide. For example, sample 2 has agglutinated with the antiserum ‘anti-B’ but not with ‘anti-A’; so this sample is defined as group A. (b) Determination of Rhesus blood groups was carried out in tubes. The still shows agglutination in a tube held up to a mirror. Researchers confirmed agglutination using a microscope. From Cyril Jenkins Productions Ltd, Blood Grouping, Imperial Chemical Industries Limited, 1955. Wellcome Library. View the film at http://catalogue.wellcome.ac.uk/record=b1750596. © ICI/Wellcome Trust, 2011. Printed with permission of Wellcome Film (Wellcome Library, London).
The Rh+ and Rh– groups soon became significant for clinicians working in blood transfusion and neonatal care; in 1941 Philip Levine, also at the Rockefeller Institute, suggested that the groups were responsible for a dangerous condition called ‘haemolytic disease of the newborn’ or erythroblastosis fetalis, in which a Rh− mother produced antibodies to her Rh+ foetus, causing the baby to be born seriously anaemic.Footnote 18 Over the next four years, the major US and British medical journals published at least eighty articles on the Rhesus groups.
‘Simple’ inheritance was already established as one of the special properties of blood groups; they were almost the only human characters whose laws of inheritance had been defined as Mendelian, and were already regarded as exemplary traits for the study of human heredity and physical anthropology, and as important tools for forensic science.Footnote 19 So after identifying the new Rhesus antisera, one of the first things that Landsteiner and Wiener did was to work towards an explanation for the inheritance of the Rh+ and Rh– antigens. They tested the blood groups of families in New York and used population frequencies to infer genetic hypotheses.Footnote 20 Their initial conclusion was that the Rhesus groups were inherited via a single gene with two variants (or ‘alleles’), one dominant to the other. Following the conventions for genetic nomenclature at the time, Landsteiner and Wiener denoted alleles using italicized two-letter symbols (Rh) and the dominance relationships between alleles using upper- and lower-case first letters (Rh and rh). Thus an individual defined as Rh+ had the genotype RhRh or Rhrh, while someone who was Rh– had the genotype rhrh. Geneticists at this time used both the terms ‘gene’ and ‘allelomorph’ to refer to variants of a gene, and the term ‘allelomorph’ both for ‘character’ variants and for gene variants; in order to avoid confusion here I use the admittedly anachronistic term ‘allele’ to mean ‘gene variant’.Footnote 21
The Galton Serum Unit in Cambridge began work on Rhesus genetics in 1942. The unit was a new incarnation of a lab that Fisher had established in 1935 at University College London with money from the Rockefeller Foundation. Fisher, together with several eminent colleagues, had, from the early 1930s, advocated the study of blood groups as a way of putting human heredity research on a genetic footing. Other human characters had ambiguous and complex inheritance, but blood groups were sharply defined traits whose population dynamics could be described mathematically.Footnote 22 For example, in his funding application to the Rockefeller Foundation, Fisher had proposed that blood groups could be developed as markers for the inheritance of other human traits, such as disease or intelligence.Footnote 23
To run his laboratory, Fisher hired serologist and former physician George Taylor, pathologist Robert Race and two other research assistants, Elizabeth Ikin and Eileen Prior. In 1939, Fisher agreed that his laboratory – the only one in the country specializing in blood-grouping techniques – should be taken over by the Medical Research Council (MRC) and made part of the new wartime Emergency Blood Transfusion Service, in which role it would make and distribute antisera to the transfusion depots around the country. When war broke out, the unit moved to the Department of Pathology in Cambridge while Fisher remained as professor of eugenics at UCL. Although they were institutionally separated, Fisher corresponded extensively with the unit – particularly with Taylor and Race – and together they made use of the vast network of blood depots around the country to obtain data and blood samples for research on the genetics of blood groups.
In 1942 Fisher suggested to his colleagues that they begin working on the Rhesus system, and soon after Taylor co-authored a national appeal in the British Medical Journal (BMJ) for doctors to send samples from mothers and babies affected by erythroblastosis fetalis: ‘Wanted: Anti-Rh Sera’.Footnote 24 Added to this, the Galton Serum Unit had strong links to blood depots and hospitals, which sent to Cambridge samples of blood from people suffering adverse reactions to transfusion.Footnote 25 The discovery of new groups was a recursive process; as the researchers tested larger numbers of blood samples, they defined new groups and antisera, and from family data they postulated new antigens, alleles and dominance relationships. As they discovered more and more antisera they struggled with patterns of agglutination and their inheritance. For the first couple of years the unit corresponded regularly with the Wiener lab, sending them new results in advance of publication. By 1944 the two labs had similar results. Despite using slightly different antisera, some of which bore different names, they had defined the alleles Rh 1, Rhy, Rh′, Rh2, Rh0, Rh″ and rh. Moreover, despite the lack of clear patterns, a single gene with several alleles was still the most promising explanation for their inheritance.
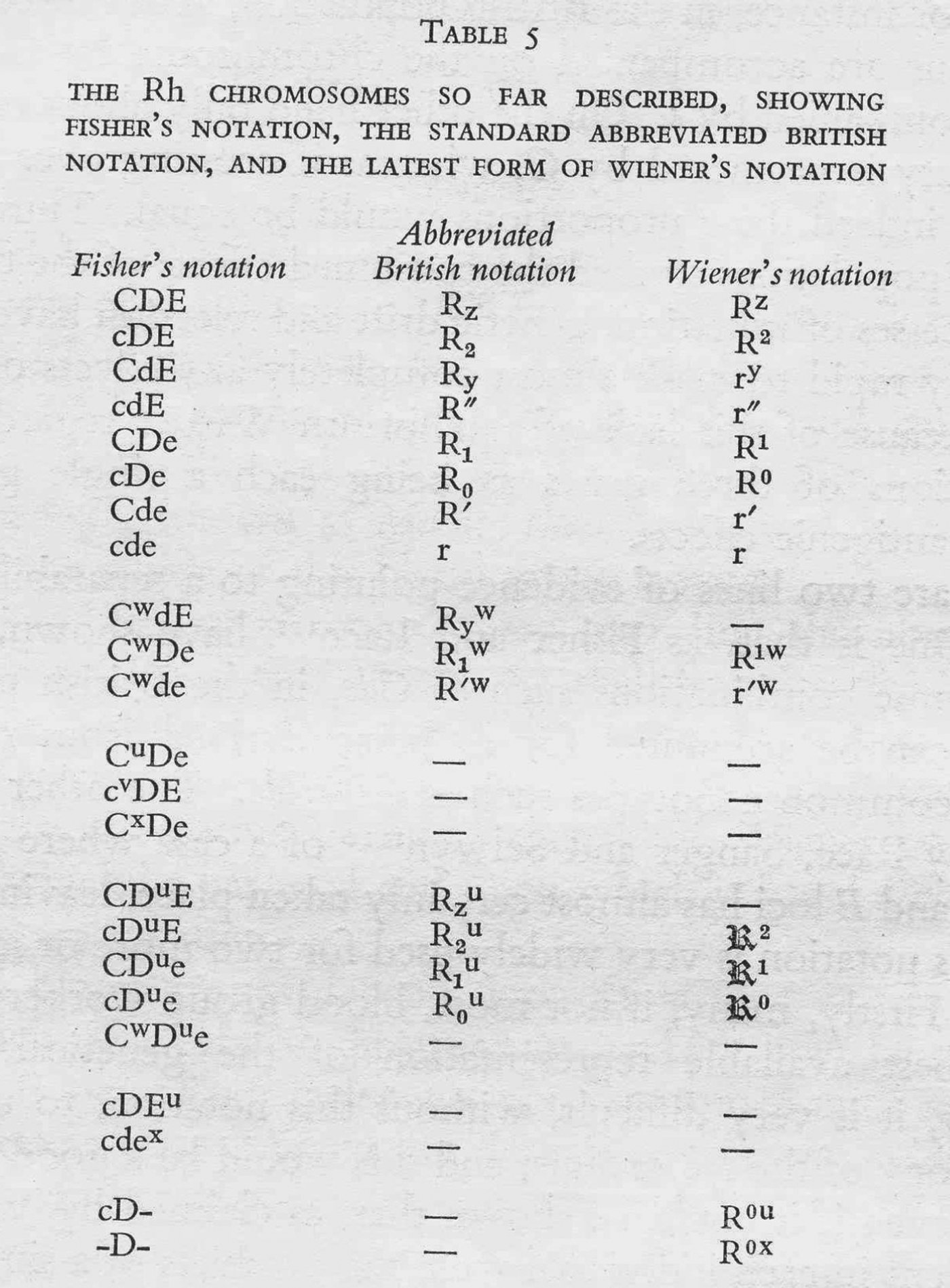
In June 1944 this transatlantic harmony was seriously disrupted by a paper in Nature in which Race described a proposal by Fisher – famously worked out in a Cambridge pub (Figure 2) – for an entirely new mechanism of Rhesus inheritance.Footnote 26 Instead of a single gene with many possible allele variants, Fisher proposed that the locus consisted of three tightly linked genes, each with two possible alleles. On the basis of this hypothesis, Race and Fisher suggested a complete revision of the nomenclatures used to denote antisera, genotypes and blood groups. They used single letters to denote the antigens and their corresponding alleles, and announced that ‘the three forms of allelomorphic antigens are arbitrarily denoted by C, c, D, d, E and e, chosen to avoid confusion with any so far used’.Footnote 27 So where the researchers had previously seen single antigens, Fisher and Race now saw combinations of three. Following a suggestion by a colleague, they also proposed new names for the antisera to reflect the antigens with which they were supposed to react: ‘anti-C’, ‘anti-D’, ‘anti-d’, etc.Footnote 28

Figure 2. Fisher reportedly first articulated his new hypothesis on a pub napkin. Image reprinted with the consent of the Fisher Memorial Trust; photograph kindly provided by Professor A.W. F. Edwards.
The Fisher–Race nomenclature spread quickly, especially in Britain, and was particularly popular among the clinicians and geneticists with whom the Galton Serum Unit collaborated. Private correspondence shows, however, that Fisher initially had to work hard to persuade even his closest allies. In a letter to a Glasgow colleague, Daniel Cappell, Fisher explained that he would have published the scheme sooner had he ‘had the real encouragement of your support at the time’, and confessed that while the theory had ‘grown on Race … Taylor still has his doubts’.Footnote 29 And indeed Fisher's triplet-gene theory was extremely bold: a tightly linked cluster of genes had never been described for humans and only once in any organism.Footnote 30 Moreover, despite initially expressing interest in the proposed system, Wiener was unhappy with the new nomenclature almost from the start, an antipathy that developed into bitterness over the next decade. Throughout this paper I use the term ‘nomenclature’, rather than ‘notation’, to denote the contested system of symbols. This is because both parties broadly agreed with the general convention that, for example, an allele should be denoted using italicized single letters or pairs of letters (e.g. Rh or C), and that dominance relationships should be denoted using lower- and upper-case letters (e.g. C and c). Thus all largely agreed on the notational system for genetics, but within those broad genetic conventions they disputed the nomenclatures appropriate for the Rhesus groups.Footnote 31
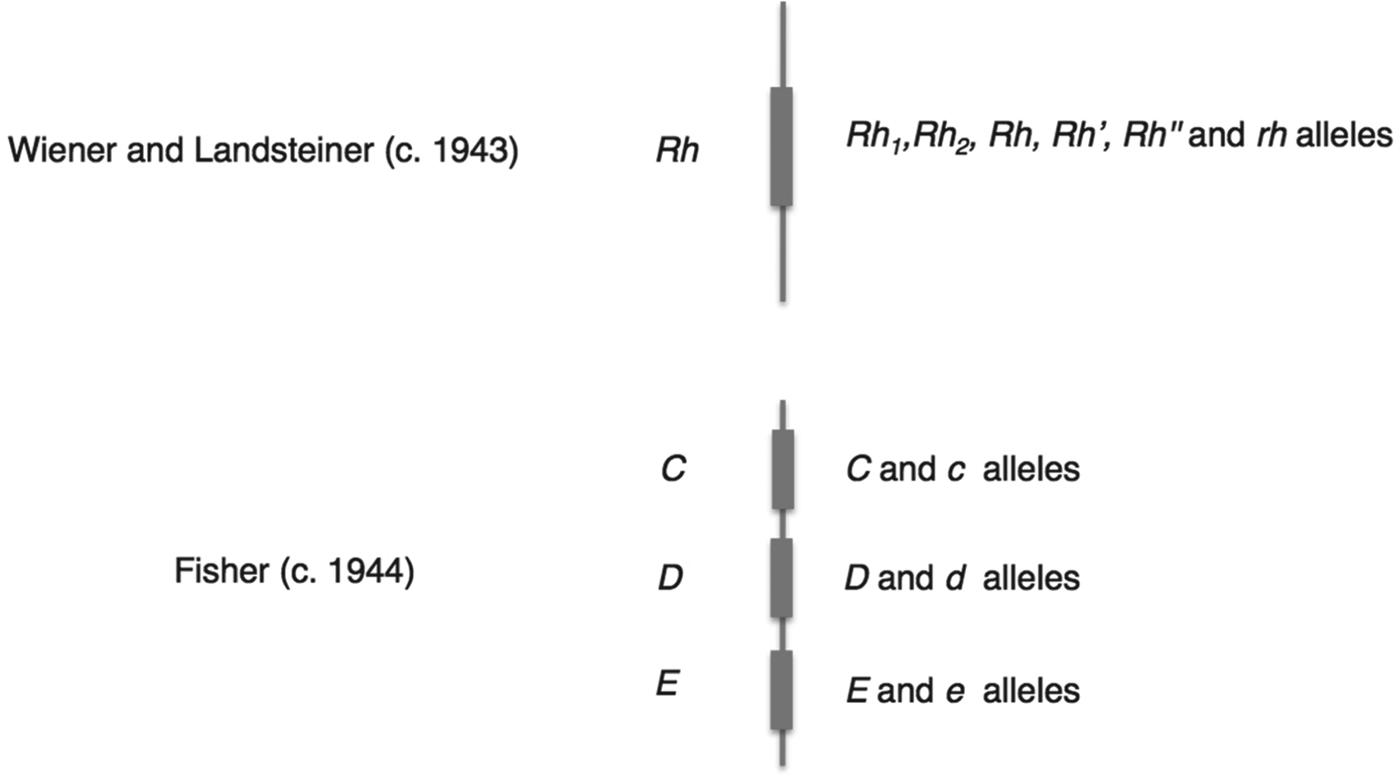
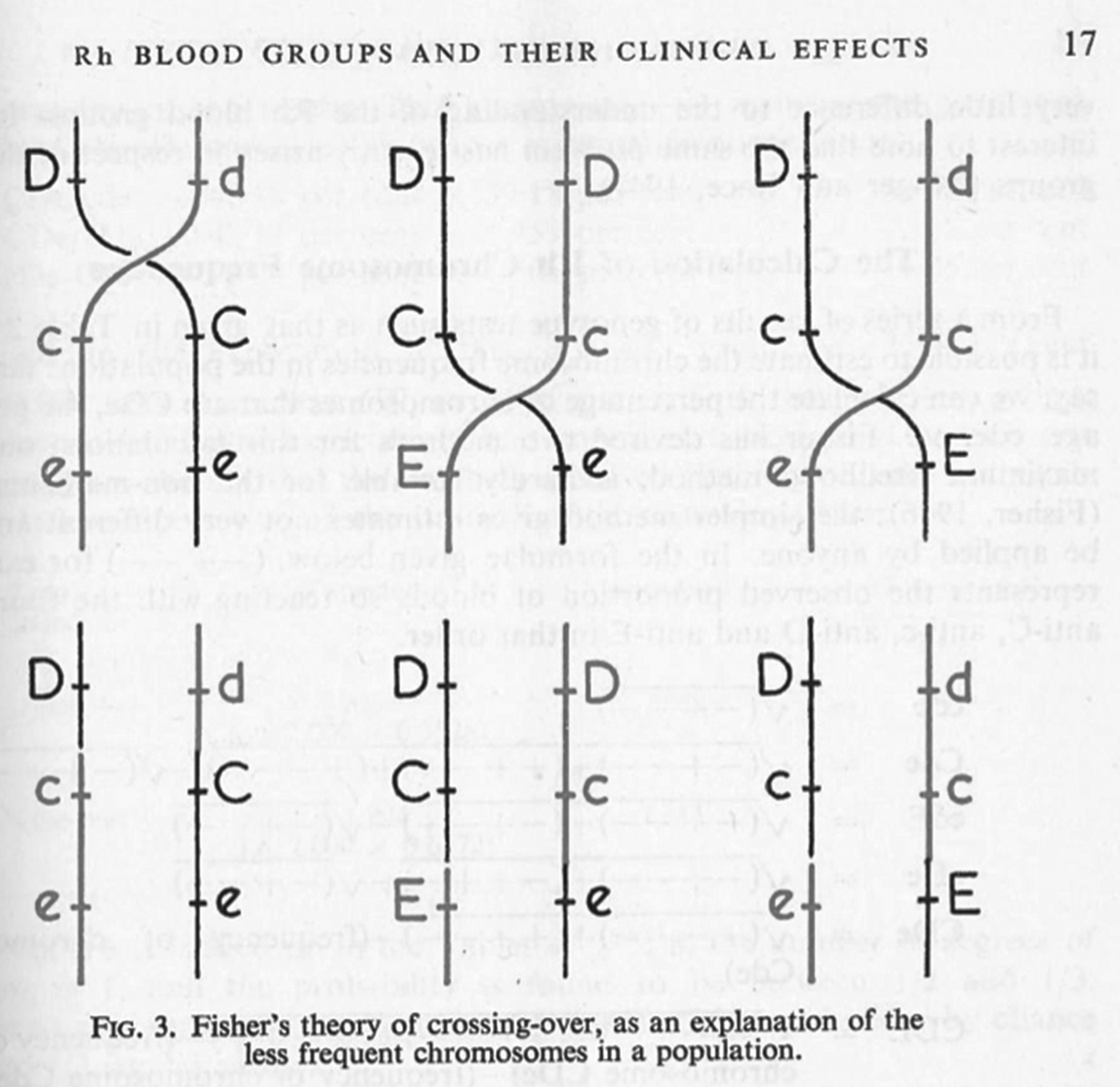
Mazumdar has explained that Wiener's and Fisher's rival nomenclatures reflected fundamentally different interpretations of how antibodies and antigens react.Footnote 32 Wiener believed that each antigen was qualitatively different, and that a single antigen – and therefore group – was defined through its reaction with a range of antisera; as he put it, ‘a single antigen molecule can react with several antibodies of different specificities’.Footnote 33 Fisher, by contrast, assumed that the interactions between antibody and antigen were all-or-nothing reactions, so that the specificity of any single reaction was generated by a triplet combination of antigens.Footnote 34 These conflicting ideas of immune specificity related to different mechanisms of inheritance. In Wiener's system, each qualitatively varying antigen corresponded to one of a range of alleles, which were all variants of a single gene (Rh 1, Rhy, Rh′, Rh2, Rh0, Rh″ and rh), while Fisher and his colleagues saw three separate loci each associated with a pair of possible alleles (see Figure 3 for a graphical comparison of the two schemes). For Fisher, the relationship between allele, antigen and blood group was direct and simple: a single allele variant corresponded to a single antigen, which reacted with a single antibody.Footnote 35

Figure 3. Schematic contrasting the genetic systems of Wiener and Fisher–Race. The genes (Rh, or C, D and E) corresponded to a number of allele variants. Wiener saw a single gene with many different possible alleles, while Fisher envisaged three closely linked genes, each with a pair of allele variants.
In summary, the Fisher–Race CDE terminology emphasized the unit-like character of the three antigens, the three separate alleles that corresponded to them, and their combinatorial specificities (CDE, CDe, CdE, Cde, cDE, cDe, cdE, and cde), while Wiener's terms suggested a series of qualitatively different alleles (Rh z, Rh1, Rhy, Rh′, Rh2, Rh0, Rh″, and rh). Moreover, the different nomenclatures referred to different objects – where Wiener still saw one allele (e.g. Rh 1), Fisher and Race saw three (e.g. CDe). In this sense the two nomenclatures defined different Rhesus systems.
For the next decade these two competing terminologies were used to denote Rhesus antisera, groups, genotypes and antigens. Between 1944 and 1948 authorities in the US and Europe organized at least three major international meetings to attempt to reach a consensus.Footnote 36 This lack of standardization also prompted scores of scientists and clinicians to write to journals to debate the virtues of the two nomenclatures and suggest new ones. It is this literature that gives us rare access to the varied practical functions of nomenclatures in the research laboratory and clinic.
Organizational tools in serology
The literature on Rhesus research reveals some of the organizational functions of the nomenclatures given both to the various serological components (for example, antisera, antigens) and to genetic ones (for example, alleles). Several analyses have shown how written protocols, registries, indexes and record cards function as tools of ‘scientific bookkeeping’ to generate, or constrain, knowledge about the natural world.Footnote 37 I suggest we can see blood groups as categories designed to organize patterns of agglutination, which took form on the laboratory bench and on paper. Take the ABO grouping test shown in Figure 1. A still from a 1950s film, it shows a simple set of reactions on a porcelain tile. To designate any one sample as A, B, O or AB, it was tested separately against both anti-A and anti-B antisera; in the case here, sample 1 reacts with anti-A but not with anti-B, and is thus designated group A and defined as containing ‘A’ antigens. Sample 3 reacts with neither anti-A nor anti-B and so is designated group ‘O’, while sample 4 reacts with both and so is group ‘AB’. The groups were in this way inferred from two-dimensional patterns of agglutination, with different antisera across the columns, and different samples down the rows. For the ABO groups shown here these patterns were simple; for the Rhesus groups, they could be very complex.
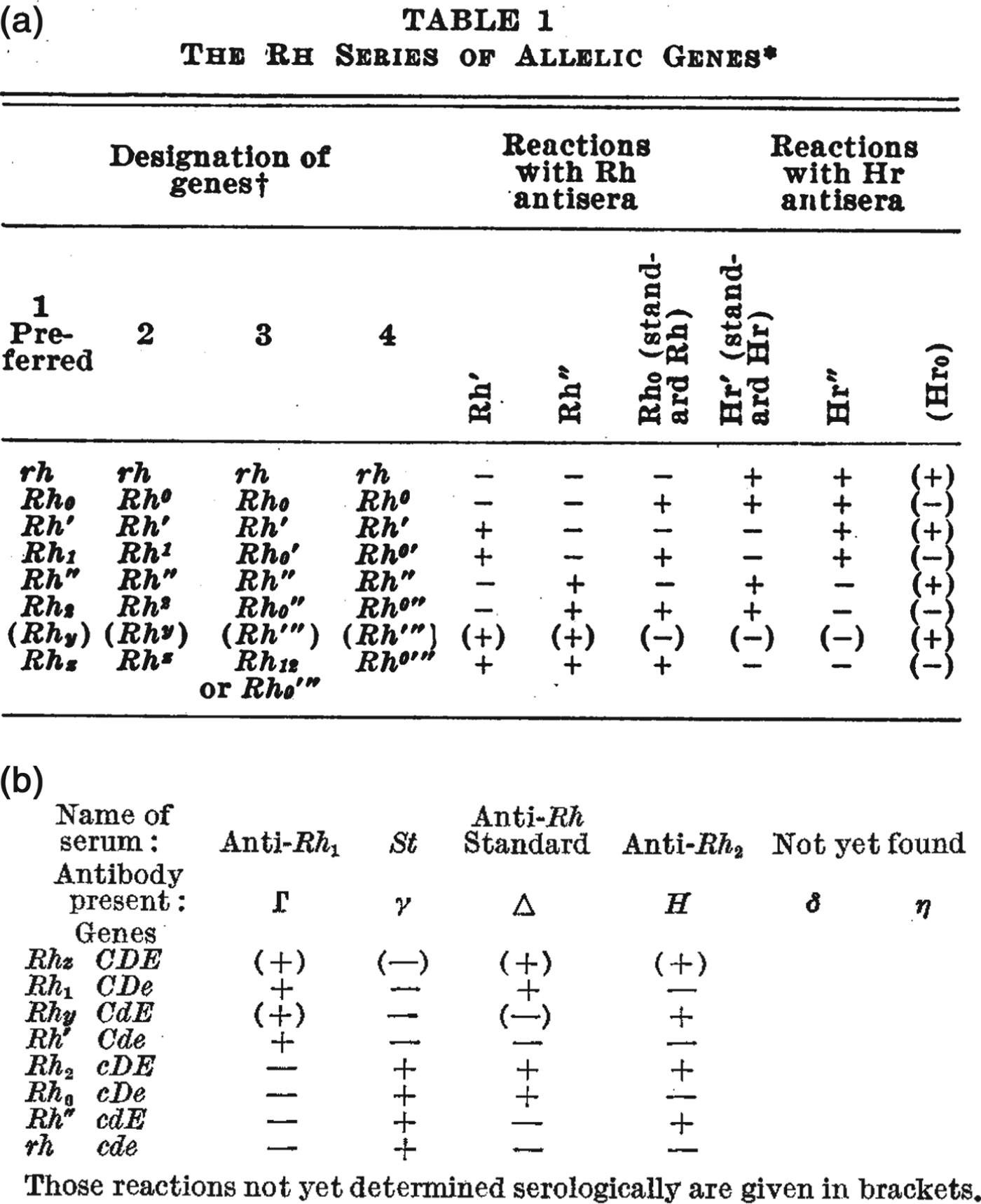
The rather more elaborate range of Rhesus tests put special emphasis on the formatting of inscriptions.Footnote 38 Researchers organized the results of agglutination reactions into two-dimensional tables – for example, the handwritten notes in Figure 2 and the published examples in Figure 4 – and often chose nomenclatures to reflect common or contrasting features of certain antisera. For example, both of the tables reproduced in Figure 4 draw attention to the observation that some antisera caused precisely the opposite pattern of reactions to others, and the nomenclatures put special emphasis on this reciprocity. Referring to the table in Figure 4a, Wiener suggested that because of their opposite agglutination reactions, the ‘anti-Rh′ serum’ might be better designated ‘simply as anti-Hr′.’ He also suggested that the antiserum with opposite reactions to anti-Rh0 might be denoted anti-Hr0.Footnote 39 Fisher and Race chose a similar strategy, but instead of reversing the letters of the symbols, they used upper- and lower-case letters to indicate serological reciprocity (Figure 4b). Even while later downplaying the significance of their controversial genetics, they drew particular attention to the importance of these serological patterns.Footnote 40

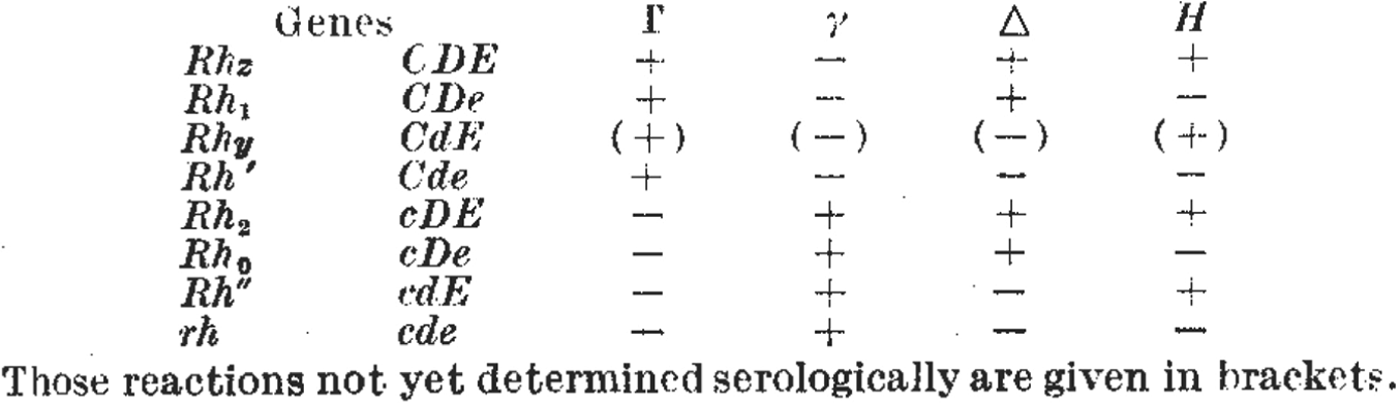
Figure 4. Figures 4a and 4b. As researchers defined new Rhesus groups they altered names and symbols to help organize the patterns of agglutination. (a) A table from a paper by Wiener in 1945 includes the new antiserum names Hr′ and Hr″. Comparing the + and – reactions along the table shows that the symbols Hr′ and Hr″ were meant to indicate reciprocal reactions to Rh′ and Rh″. From Wiener, ‘Theory and nomenclature of the Hr Blood Factors’, Science (1945) 102, pp. 479–482. Reprinted with permission from AAAS. (b) Table from the paper by Race that first described Fisher's new hypothesis. Here, the reciprocal allele names (C and c, D and d, E and e) were chosen to reflect contrasting agglutination reactions. For example, look vertically down the table to compare the + and – reactions for antisera Γ and γ corresponding to the putative alleles C and c. From R.R. Race, ‘An “incomplete” antibody in human serum’, Nature (1944) 153, pp. 771–772. Reprinted with permission of Nature Publishing Group, www.nature.com.
For Fisher and Race, their new theory and its nomenclature also had important predictive functions. First, the reciprocal reactions of the antisera Γ (later ‘anti-C’) to γ (‘anti-c’) suggested the existence of antithetical sera to Δ (‘anti-D’) and Η (‘anti E’).Footnote 41 Sure enough, η (later ‘anti-e’) was discovered by a young doctor volunteering at the Luton blood depot, Arthur Mourant, who very soon joined the Galton Serum Unit himself.Footnote 42 Second, the theory and its nomenclature predicted an eighth allele that had not yet been defined (CDE or Rh z), but which was found soon after (compare the first rows in Figures 4b and 5). The researchers had been able to make this prediction owing to the combinatorial specificity of Fisher's theory, and because its symbols could be combined and recombined in tables and grids.

Figure 5. Table from Murray, Race and Taylor (1945), which described identification of the new Rh z gene and its predicted (Figure 4b), now confirmed, serological reactions. From R.R. Race, G.L. Taylor and John Murray, ‘Serological reactions caused by the rare human gene Rhz’, Nature (1945) 155, pp. 112–114. Reprinted with permission of Nature Publishing Group, www.nature.com.
This combinatorial feature of the Fisher–Race system was important in another way too. The idea that the three genes (C, D and E) were tightly, but not completely, linked corresponded to the prediction that from generation to generation they might, albeit rarely, form new combinations. This, Fisher argued, explained why, among the general population, some CDE combinations were common, some very rare and others extremely rare (Figure 6). Since many geneticists were eager to establish linkage in humans, they found the idea of tightly linked alleles appealing, and it was a hypothesis that could be tested by studying large populations. The Galton Serum Unit's links to blood depots and hospitals did eventually yield enough data to provide the very rare recombination events that the theory predicted.

Figure 6. Fisher suggested that recombination could occasionally generate very rare but predictable combinations of alleles (bottom row) from the more common types (top row). Figure from P.L. Mollison, A.E. Mourant and R.R. Race, ‘Medical Research Council Memorandum no. 19: The Rh Blood Groups and their Clinical Effects’, His Majesty's Stationery Office, London, 1948. Made available for republication under the Open Government License, www.nationalarchives.gov.uk/doc/open-government-licence.
Both groups of researchers struggled to find nomenclatures that made sense of patterns of agglutination. But not only was Fisher's bold new proposal predicated on a different conception of how antibody reactions worked; he and his colleagues argued that it also made better sense of Rhesus inheritance. I now want to look more closely at what it meant to organize patterns of inheritance on paper, particularly for the practice of ‘working out’ genetic crosses.
Nomenclature as genetic objects
Studying heredity requires keeping track of characters across several generations and so puts particular emphasis on the organizational functions of paperwork.Footnote 43 Wiener, Fisher and Race all attempted to infer the genetics of the Rhesus system by recording the phenotypes of family members belonging to several generations. The researchers transposed this data into phenotype frequencies, collated these in tables, and tried out plausible genetic crosses. ‘Working out’ crosses was an experimental practice; researchers would try out putative genotypes, and use them to generate expected phenotype frequencies that could then be compared with observed frequencies. Having established a genetic hypothesis, researchers would use it to infer possible genotypes and calculate allele frequencies.
In this way, nomenclatures functioned to define genes, alleles, their dominance relationships and their rules of inheritance.Footnote 44 For both the Wiener and Fisher–Race nomenclatures the one-to-one correspondence between symbol and allele (and antigen and antibody) lent the letters a ‘graphic suggestiveness’ that was significant for their deployment in genetic crosses.Footnote 45 Each symbol denoted an indivisible, discrete object. Where Wiener saw a single allele, Fisher and his colleagues saw three. Genes and alleles were symbols that organized observations about inheritance. They were also the materials that the researchers had to work with, and constituted objects of research.
Those involved in the Rhesus dispute sometimes made allusions to the algebraic qualities of genetic nomenclatures. The British workers referred to the CDE symbols as ‘antigenic formulae’ that could predict blood types and antisera that had not yet been defined.Footnote 46 In Science, two clinical geneticists in Michigan, Edward Ducey and Robert Modica, both committed to the Fisher–Race hypothesis, complained that using Wiener's nomenclature ‘reminds one of multiplying with Roman numerals, i.e., XIV x VIII = CXII! … One must memorize the entire table; one cannot easily work out the genotypes involved’.Footnote 47 Ducey and Modica implied that complex transpositions were needed to make the two systems commensurate. Wiener's nomenclature could not easily be used to predict dominance relationships of the Fisher–Race system because his symbols defined different genetic objects.
But researchers carried out a range of different kinds of calculation, and the notebook scribblings from Robert Race's laboratory show that, like laboratory instruments, nomenclatures were good for some kinds of calculation and not others. Sometimes a single nomenclature was not sufficient for different kinds of work. The 1948 MRC Memorandum on the Rhesus groups, written by Race, Arthur Mourant and their haematologist colleague Patrick Mollison, introduced a range of ‘short symbols’ for the Rhesus genotypes, most of which were ‘based on those by Dr A.S. Wiener’.Footnote 48 For example, ‘cde/cde’ was given the shorthand ‘rr’, and ‘cDE/cdE’ was reformulated as ‘R 2R″’.Footnote 49 These symbols were so useful that Race and Sanger included them in the first edition of their textbook Blood Groups in Man (1950), describing them as ‘necessary’ and ‘much used’ (see also Mourant's table in Figure 7).Footnote 50 And Race and his colleagues used these shorthand symbols not only in printed text but also in private laboratory scribblings. In the lower section of the laboratory notes shown in Figure 8, Race worked out the distance between the genes C, D and E using genotype frequencies within families.Footnote 51 But in the separate calculations in the top part of the page, he denoted genotypes using the shorthand symbols. So even on a single piece of paper, different kinds of Rhesus notation were deemed appropriate for different contexts and functions.

Figure 7. The Wiener-modified shorthand (or ‘Abbreviated British notation’), from A.E. Mourant, The Distribution of the Human Blood Groups, Oxford: Blackwell Scientific Publications, 1954, p. 14. All reasonable attempts have been made to contact the copyright holders of this image, and it is reprinted here with the consent of the Mourant estate.

Figure 8. Piece of paper showing Robert Race's calculations and his deployment of both the CDE nomenclature and the Wiener-modified shorthand nomenclature. From Race, ‘Blood Group Research Unit papers’, 7 February 1947, SA/BGU/C.1, Wellcome Library. Reproduced with permission from the Medical Research Council.
Symbols had a special material significance in genetics. Genes could not be seen directly; they were inferred from family and population data and represented as symbols on paper. These symbols were objects to work with – sometimes transitory, existing only during the course of an experiment, and sometimes deployed in more durable settings, such as in scientific papers or textbooks.
Oral, handwritten and typographical articulation
To understand more fully the ways that nomenclatures shape and are shaped by their functions, I now follow the Rhesus blood groups out of the laboratory notebook and into other settings and material forms. Different nomenclatures presented different kinds of typographical challenge depending on whether they were made using typewriters or printing presses, or by hand. During an attempt to standardize the names of Rhesus antisera on bottles, a committee convened by the US National Institutes for Health (NIH) noted that the Fisher–Race nomenclature was easier to handle typographically than Wiener's, which involved ‘complications, both typographical and genetic, of subscripts, superscripts, numbers, primes, and other symbols’.Footnote 52 The NIH committee declared that the Fisher–Race nomenclature was, by contrast, ‘simple and direct, both typographically and genetically’.Footnote 53 The implication was that although subscripts, superscripts and primes might be relatively easy to deploy in handwriting, they presented greater problems for the typewriter and printing press.Footnote 54
But handwriting on paper and blackboard also presented important challenges. Zoologist and human geneticist Herluf Strandskov remarked that while the Fisher–Race nomenclature was easy to deploy in notes, the Wiener terminology was ‘not easy to present clearly when written in longhand on paper or on the blackboard’.Footnote 55 Meanwhile, the instability of handwriting meant that the upper-case ‘C’ and lower-case ‘c’ might generate dangerous ambiguities, especially if lower and upper case were not displayed together.Footnote 56 Ducey and Modica, who supported Fisher's triplet-gene theory, proposed a modified version of the Fisher–Race nomenclature whereby the names of the antisera included only lower-case letters. Ducey and Modica suggested removing capital letters from the antisera, reserving them instead for the antigens, and replacing the upper-/lower-case alleles with a system of primes. This, they maintained, would preserve the reciprocal relations between antigen and antiserum, but preclude confusion in the ‘reading of these terms’.Footnote 57
Meanwhile, the Rhesus nomenclatures also had to be articulated orally, in the genetics laboratory, the clinical research laboratory and the hospital. Ducey and Modica argued that their version of the Fisher–Race nomenclature would prevent confusion in speech. Elsewhere, researchers complained that the Fisher–Race system's dependency on upper- and lower-case alleles made the spoken articulation of genotypes extremely cumbersome. The antisera names were not too difficult; in a British film about grouping technique released in 1955, the narrator simply refers to ‘big D’ antiserum.Footnote 58 But genotypes were problematic, since the single genotype cde/CdE would have to be said as ‘little-c, little-d, little-e over big-C, little-d, big-E’. Dallas haematologists Joseph Hill and Sol Haberman, working in a clinical pathology lab and hospital blood bank respectively, declared, ‘One of the most frequent criticisms that have been made of the Fisher–Race notation is that it is difficult to use verbally.’Footnote 59 They suggested a whole new system for speaking the nomenclature that, they claimed, would be ‘useful to the laboratory worker as well as to the clinician’. This involved articulating only the upper-case allele, and preceding it with the terms ‘homozygous’ and ‘heterozygous’; it meant pronouncing the genotype ‘Cde/CDe’ as ‘homozygous C, heterozygous D’, leaving the homozygous ‘e’ silent.Footnote 60
Spoken articulation was another reason why even Fisher's closest supporters felt that a version of Wiener's nomenclature could function as a useful shorthand. In Blood Groups in Man (1950) Race and Sanger explained that the shorthand symbols were ‘convenient in conversation’ and ‘desirable when long lists are being drawn up’.Footnote 61 The problem of spoken articulation was also dealt with by textbooks. Following general conventions in genetics, both nomenclatures used italicized letters to distinguish alleles from their corresponding antigens; the American textbook Blood Transfusion (1949) gave precise instructions for how to make that distinction without the italicization available to printed inscriptions. Describing Wiener's nomenclatures, for example, it declared that ‘in vocalizing, the [genotypes] are distinguished from the type names by the lack of h's in the gene characters’, and it advised its readers that on paper it was best to drop the ‘h’ from the Rh symbols.Footnote 62 Speech exerted significant pressures on nomenclatures, and considerable effort was made to facilitate and standardize their articulation.
In summary, a controversy precipitated by a theoretical dispute about genetics and immunology brings to light a more complex ecology of laboratory and clinic, where different nomenclatures not only moved between people with different interests at stake, but took on various material forms. We see that the nomenclatures were deployed using printing presses and typewriters and through handwriting, but were also applied to the blackboard and articulated in speech. This range of forms gives us a new possibility for why the Rhesus controversy was so prolonged. Numerous letters and articles suggest that different nomenclatures had different virtues in different settings, and that no single nomenclature was sufficiently flexible for all of these contexts.
Nomenclatures privilege different objects
We have seen that blood samples, antisera and data records were circulated between laboratories and hospitals, and among geneticists, immunologists, clinical pathologists, blood bankers and anthropologists. Now I want to consider ways in which researchers in these different settings also tried to make nomenclatures reflect commitments to certain practices.
An advocate of the Fisher–Race system, Glasgow physician Daniel Cappel, argued that antisera should reflect the names of the antigens and their alleles. The earliest version of the Fisher–Race nomenclature had denoted the antisera using Greek letters (such as Δ, Η, Γ), as was customary for antibodies more generally (Figure 4b). But Cappell soon suggested that the antisera be renamed ‘according to the elementary antigens of the Rh complex with which they react’, so that antisera would be called ‘anti-C’, ‘anti-D’ and ‘anti-d’ etc. He argued that this new system had ‘the further merit that it is easily adaptable as knowledge advances’, ‘knowledge’ clearly meaning genetic knowledge.Footnote 63 This terminology, privileging reference to the allele names, was rapidly accepted and developed by Race, Fisher and their geneticist colleagues.Footnote 64
Conversely, others argued that the genotype terminology should reflect the actions of the antisera. In 1944, John Murray, an assistant pathologist at the Middlesex Hospital and collaborator with Fisher and Race, suggested in Nature that the Rhesus antisera should be assigned the numbers 1, 2, 3 and 4, and that the genotypes could then be given names that reflected the patterns of agglutination with those antisera. This, he explained, would mean that ‘at a glance it may be seen exactly with what sera the cells have been tested’.Footnote 65 In Murray's scheme, various antigens would therefore be denoted Rh 136, Rh 126, Rh 123 etc., with supernumerals indicating the sera with which the antigens reacted. So although Murray accepted the 1:1:1 mapping between antibody, antigen and allele, his nomenclature privileged the clinical significance of antisera, which were likely to have been immediately significant to clinical pathologists trained in serological techniques.
Meanwhile, a doctor from Birmingham, Henry Baar, also suggested a nomenclature in which antisera reactions were reflected in the blood-group names. His rather eccentric proposal was a hybrid of both the Wiener and Fisher–Race systems. Baar proposed ‘that the phenotype be designated by the letters of the sera used in testing added to the statement Rh+’. It resulted in the very unusual system shown in Figure 9; perhaps owing to the great difficulties in handling this typographically, it appears nowhere else.Footnote 66

Figure 9. Section from paper by Baar illustrating his suggested nomenclature for the Rhesus genes. H.S. Baar, ‘Anti-Rh serum nomenclature’, British Medical Journal (1948) 4562, 1156–1157. Reprinted with permission from BMJ Publishing Group Ltd.
A particularly puzzling feature of the Rhesus controversy is why the official guidelines for labelling bottles of antisera never resolved the dispute one way or the other. For example, the conveners of the 1948 NIH meeting reluctantly admitted in the journal Science that they had been ‘forced to the conclusion that for the present a compromise must be made’, recommending that ‘the Wiener terminology appear first … followed by the Fisher–Race terminology in parentheses’.Footnote 67 This ruling – upheld in 1953 by the Food and Drug Administration – has often been seen as a failure to resolve an urgent problem.Footnote 68 But we might ask why there was not more pressure from clinicians to standardize the labelling of antisera.
Part of the answer may have to do with the fact that usually only two of the Rhesus groups, Rh+ and Rh–, caused incompatibility with any serious medical effects. These groups were distinguished simply using the antiserum called ‘Rh0’ by Wiener and ‘anti-D’ by Fisher and Race.Footnote 69 After 1942, when Rhesus inheritance first became important for erythroblastosis fetalis prognosis, clinicians were kept abreast of genetic research on the system by books and medical journals. But by the end of that decade, medical textbooks often emphasized that for clinicians the complex genetics of the system was largely irrelevant. The British textbook Blood Transfusion (1949) explained Rhesus genetics in detail, but its picture of a ‘specimen label attached to blood bottles’ indicated only the simple Rh+/Rh– denotation (Figure 10).Footnote 70 The BMJ reminded its readers that despite the exciting nomenclatural controversies at stake, ‘only one, D (big D), is of great clinical importance’.Footnote 71 Textbooks such as Mollison's Blood Transfusion for Clinical Medicine (1951) carefully explained the Fisher–Race genetics but then stated that ‘fortunately … a simple subdivision of human beings into Rh positive and Rh negative … is sufficient for routine clinical purposes’.Footnote 72 Though he was a close and sympathetic colleague of Race, Mollison declared in a BMJ review of Blood Groups in Man (1950) that it was ‘useless to pretend any longer that what is interesting and even fundamental in the field of blood groups is necessarily of the slightest interest in clinical medicine’.Footnote 73 So in many ways the Rhesus system was relatively simple for clinicians, perhaps resulting in little pressure to resolve the nomenclature one way or another.

Figure 10. Labels attached to bottles of blood for transfusion, from H.F. Brewer and G. Keynes, Blood Transfusion, Bristol: John Wright, 1949, p. 385. All reasonable attempts have been made to contact the copyright holders of this image.
Indeed, a crucial oddity of this story is why a debate relevant to a relatively small community of researchers drew in so many clinicians, pathologists and serologists. We have seen that the controversy began as a dispute about immunology and its genetics, and of course these had variable importance for workers in other settings, depending on their training. But an additional explanation for the controversy's long reach is the practical services offered by the Wiener lab and the Galton Serum Unit among the network of communities engaged in blood grouping. During the Second World War the unit was the primary institution for the production and circulation of standardized antisera to transfusion centres, as well as a centre of expertise for difficult grouping problems. Meanwhile Wiener – as head of the Blood Transfusion Division at the Jewish Hospital in Brooklyn and professor at the Department of Forensic Medicine at New York University School of Medicine – also circulated antisera between laboratories of different kinds; by the end of the 1940s he had a licence from the US National Institutes for Health to make and distribute antisera to labs and hospitals. Thus transfusion and clinical communities depended on reactions circulated by these laboratories, which were in turn able to exert considerable control over the ways in which the Rhesus groups were interpreted.
To summarize, researchers often tried to make blood group nomenclatures match particular specific aspects of blood-grouping practice, or certain serological or genetic features of the system. But it was hard to find a single satisfying solution that could be comfortably shared between such diverse settings and used for the full range of purposes. In the first edition of their textbook (1950), Race and Sanger attempted to deal with this problem by including a section on ‘Rh notation’, which listed the nomenclatures appropriate for different settings. Under the headings ‘clinical purposes’ and ‘laboratory purposes’, the authors stipulated those nomenclatures that would variously cause ‘the minimum of alarm’, were the most ‘convenient’, or best represented immunological understanding.Footnote 74 Users had to settle for a sanctioned diversity of terminologies.
The controversy fades
By the mid-1950s Fisher's triplet-gene theory had become generally established as the more accurate description of the genetics of the Rhesus system, although its predicted anti-d serum never materialized. The broad acceptance of the Fisher–Race nomenclature was partly due to the publishing success of its key advocates. For example, Race, Mourant and Mollison authored several MRC memoranda on Rhesus grouping, while Race and Sanger's book Blood Groups in Man (1950) eventually ran to six editions, becoming a standard textbook on blood grouping for clinicians, geneticists and anthropologists.Footnote 75 By the mid-1950s, the Fisher–Race nomenclature had largely (but by no means wholly) won out, and editors had ceased to publish Wiener's diatribes against it.Footnote 76
Nevertheless, within another ten years the Fisher–Race nomenclature itself was something of a relic. By the fifth edition of Blood Groups in Man (1968) the clear symmetry of the CDE system had been complicated by thirty Rhesus antisera and over forty tightly linked gene complexes. Sanger and Race reflected on these problems, admitting that ‘neither the CDE nor Dr. Wiener's Rh–Hr notations have found it easy to digest the surfeit of more recent complex antibodies and antigens’.Footnote 77 But they explained that they still felt that theirs was the ‘only notation in which a detailed account can be communicated’.Footnote 78 Nevertheless they wondered whether the CDE nomenclature might impede progress in the field, noting that ‘for further advance [it] must be freed of all interpretive meaning’. They also suggested that in some circumstances the Rhesus system might ‘be stored more efficiently by the numerical notation’, a terminology that was first proposed in 1961 at a meeting of the American Association of Blood Banks. Race and Sanger finished, rather wearily, with the declaration that the CDE nomenclature had to be ‘thawed out and reconstituted in familiar terms, at any rate for the present generation of workers’.Footnote 79 Nomenclatures were once again destabilized by changes to the Rhesus system.
However, as before, the explanatory virtues of a notation had to be balanced against its practical uses, demands that were sometimes, although not always, conflicting. The ‘numerical terminology’ alluded to by Race and Sanger was established to denote some of the newer kinds of specificity that older systems could not accommodate, but Mazumdar notes that in the end the numerical system was only used for punch-card or computerized data collections. Once again, then, a single nomenclature was only suitable for a limited range of media. Indeed, the CDE terminology itself never quite went away. Even today, the standard Rhesus antiserum is called ‘anti-D’, we have ‘RHCE’ alleles that correspond to ‘RhCE antigens’, and a combination of alleles might be referred to as the ‘cde haplotype’, ‘CDe haplotype’ and so on.Footnote 80 So although the genetics of the Rhesus system outgrew the CDE nomenclature, it remained useful enough to be retained in some contexts.
Conclusion
I suggest that the Rhesus dispute was prolonged by the diverse contexts and material forms in which its nomenclatures were expressed. The literature generated by the controversy brings into view the ways that researchers deployed nomenclatures as working objects for organizing serological results and genetic experiments on paper, similar to the functions of chemical symbols and formulae. It also shows us the different ways that nomenclatures were articulated in the quotidian media of genetic research. I have argued that notebook jottings, typewritten letters, published papers and speech all constituted different material forms for inscriptions, and no single nomenclatural system was sufficiently flexible to work in all settings. Even on a single piece of paper, a variety of nomenclatures might be deployed in different kinds of calculation and note-taking. Moreover, a symbol that was easy to write on a blackboard was not always so easy to say.
Another possible reason why the Rhesus nomenclatures were so troublesome was that they were passed between people working in diverse disciplines. Extending existing analyses of the ways that diagrams and models function in new domains, I have suggested that their circulation can be problematic even for the most mundane and abundant nomenclatures. Blood-group nomenclatures were generally difficult to negotiate because of their complex status and meanings in so many different settings. Almost as soon as the Rhesus groups were discovered they were eagerly taken up by geneticists, immunologists and doctors, and later anthropologists and forensic scientists. Users had loyalties to different practices and objects, and while geneticists defined more biochemical types, doctors and transfusion workers deliberately retained simpler nomenclatures in the clinic. By the late 1940s, textbooks even stipulated which nomenclatures were appropriate for which settings.
As the Rhesus controversy suggests, human geneticists occupied a particularly interesting disciplinary position, as they exerted authority over some objects of research but also relied on materials and data collected by researchers in other fields with different priorities. The consolidation of human genetics began in earnest in the 1940s and coalesced in the late 1950s, which saw, for example, the First International Congress of Human Genetics.Footnote 81 During this period, several researchers called for blood-group nomenclatures to be brought into line with the systems used in other subfields of genetics. In 1948, human geneticist Herluf Strandskov of the University of Chicago lamented that ‘no uniformity and no conformity to established rules’ existed for blood-group nomenclature. He complained that this had led to ‘confusion and to the impression … that little or no exact human genetic knowledge is at hand, when actually much precise information is available’.Footnote 82 E.B. (Henry) Ford, a geneticist friend of Fisher's at the University of Oxford, also argued that ‘confused terminology’ had ‘been a potent factor in preventing many geneticists from including the blood groups within their sphere of interest’.Footnote 83 In a private letter he asked Race, ‘Why, my dear Rob, does this still go on? General geneticists outside blood grouping cannot understand it, and it prevents them taking an interest in serology.’Footnote 84 We might generally expect to see particularly intense efforts to change and standardize notations when practitioners are attempting to align fields and subfields.Footnote 85 Strandskov and Ford wanted to bring the study of human inheritance into line with the rest of genetics and believed that the nomenclatures were putting geneticists off studying the topic.
So following the negotiations of nomenclatures might help to access the communities, practices and authorities of genetics. Today, the discipline is large and heterogeneous enough for different genetic communities to maintain different terminological conventions and standards. In plant and animal genetics these were for many years stipulated by the 1998 Trends in Genetics Genetic Nomenclature Guide; now journal editors increasingly rely on community websites.Footnote 86 Over and above that, even for a single organism, consensus needs to be negotiated for clusters of genes with related functions.Footnote 87
Nomenclatures are faced with perpetually competing pressures to be, on the one hand, finely tuned tools for particular jobs, and, on the other, objects to be circulated within and between communities. Genetic symbols can be transient and local, and exist only in the course of some experimental scribbling on paper, or made more durable through journal publications or textbooks. I have suggested that for Fisher and Race, symbols were the only ways in which genes were materially circumscribed and manipulated – they could not be seen directly, but were constructed on paper from phenotype frequencies. Today, the Human Gene Nomenclature Committee (established in 1979) aims to ‘strike a compromise between the convenience and simplicity required for the everyday use of human gene nomenclature and the need for adequate definition of the concepts involved’.Footnote 88 In genetics, nomenclatures have practical functions, but also define concepts. One reason why Robert Race resisted Ford's plea for nomenclatural standardization in the mid-1950s was that he felt it would close down too quickly complexities that were still emerging, the implication being that nomenclatures could fix understanding. Race articulated this in a letter to a colleague in the US, writing, ‘I think our knowledge of blood groups and of the nature of the gene is growing, or perhaps shrinking, so rapidly that any sort of laying down the law about notation would be a nuisance – whichever way a decision went.’Footnote 89
Moreover, the entity ‘gene’ has changed in quality and meaning throughout the twentieth century.Footnote 90 Gene concepts have shifted as new kinds of genetic object are defined. After all, the protein-coding portions of genes no longer represent the only genetic objects studied – we have, for example, retroelements, SNPs, microsatellites, SINES and LINES, and elements that influence gene expression; the nomenclatures of all of these are expected to reflect taxonomic and functional characteristics.Footnote 91 Moreover, where geneticists in the mid-twentieth century defined and manipulated their objects of research on paper, many modern researchers interact with sequence data through, for example, the online Basic Local Alignment Tool (BLAST).Footnote 92 Nomenclatures are repeatedly interrogated and contested as the relationships between genetic subfields shift.Footnote 93 Such changes might be traced in the terminology and symbols used to define genes and other genetic objects. Following nomenclatures will help to recover the ways in which geneticists visualize and articulate their objects of research, how they interact with and rely upon laboratory instruments, and how they communicate and collaborate with scientists in other fields.