Once formed, the chemical bond network of the protein backbone remains highly stable during its lifetime. Very few spontaneous reactions are known to threaten its integrity; the deamidation prompted isomerization of Asn and Asp (and to a lesser extent of Gln and Glu) residues is one, leading to changes both in the constitution (αAsn→αAsp/βAsp and αAsp→αAsp/βAsp (aspartate/isoaspartate)) and stereochemistry (l-Asn→l-Asp/d-Asp). Completed via a succinimide ring formation and subsequent hydrolysis (Fig. 1), isomerization with bond-rearrangement occurs, in some cases within hours. The change in protein sequence and especially the insertion of a –CH2– group into the backbone (αAsn→βAsp) can alter its three-dimensional (3D)-structure and thus, its bioactivity profile (Uchida and Shibata, Reference Uchida and Shibata1981): for example correlation was established between the appearance of isoaspartate and structural changes of amyloid β of Alzheimer's disease (Shimizu et al., Reference Shimizu, Watanabe, Ogawara, Mori and Shirasawa2000), or in the Cu, Zn binding of superoxide dismutase (SOD1; D’Angelo et al., Reference D'Angelo, Trojsi, Salvatore, Daniele, Raimo, Galletti and Monsurrò2013; Shi et al., Reference Shi, Rhodes, Abdolvahabi, Kohn, Cook, Marti and Shaw2013) resulting in amyotrophic lateral sclerosis (for further examples see: Supplementary Table 1). Though modulated by chemical, sterical, and sequential effects (Tyler-Cross and Schirch, Reference Tyler-Cross and Schirch1991; Kosky et al., Reference Kosky, Razzaq, Treuheit and Brems1999, Reference Kosky, Dharmavaram, Ratnaswamy and Manning2009; Goolcharran et al., Reference Goolcharran, Stauffer, Cleland and Borchardt2000; Xie et al., Reference Xie, Aubé, Borchardt, Morton, Topp, Vander Velde and Schowen2000; Li et al., Reference Li, Gorman, Moore, Williams, Schowen, Topp and Borchardt2005), the conversion is not restricted to a consensus sequence motif, thus in principle, any Asn or Asp can isomerize (Truscott et al., Reference Truscott, Schey and Friedrich2016). The prevalence of the most sensitive –AsnGly– or –AspGly– sites is about 0.30–0.35% in the human proteome and thus, three of these ‘Achilles' heels’ are likely to occur in approximately every 1000 residues.
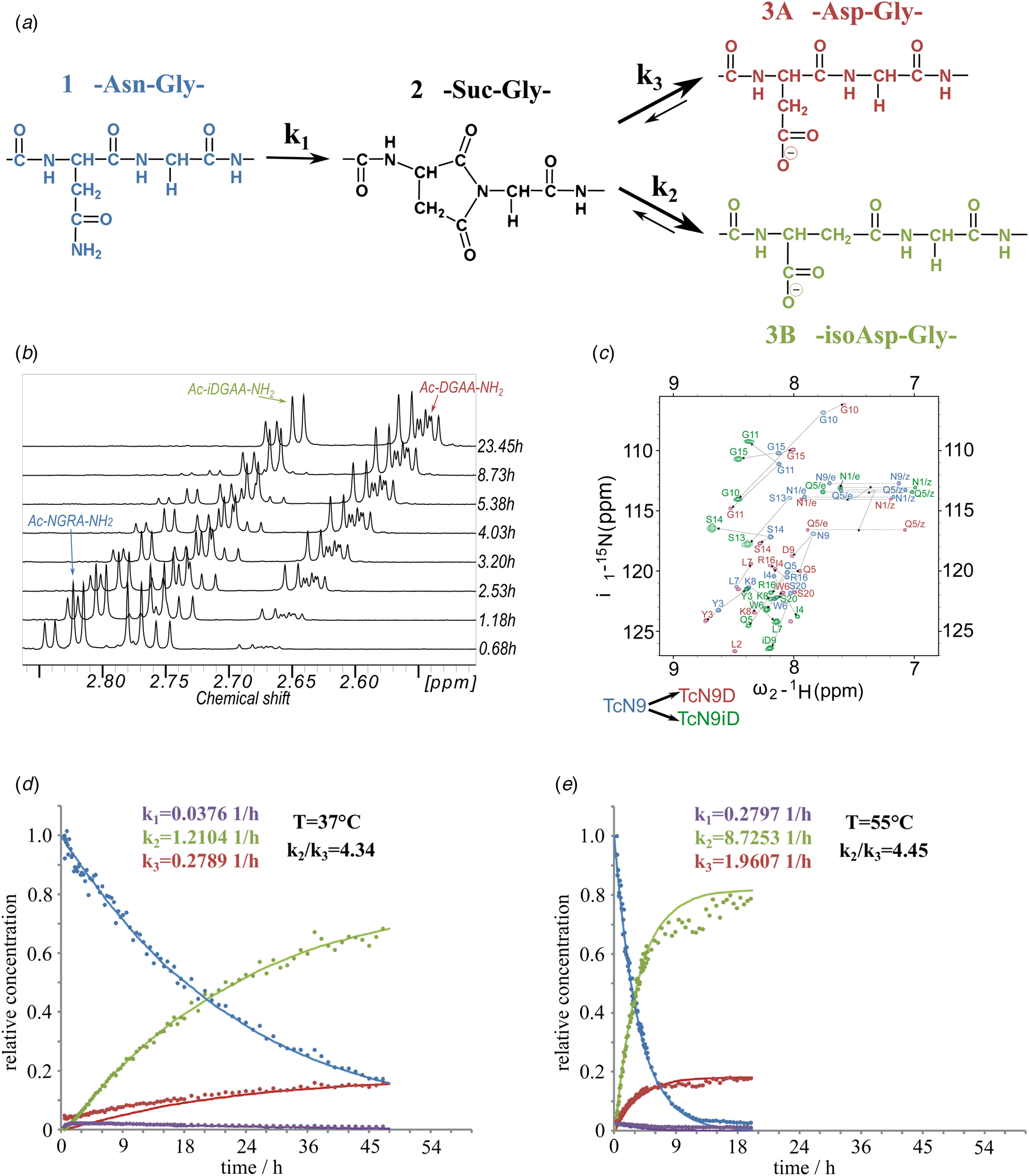
Fig. 1. (a) Isomerization reaction and rate constants of Ac-NGAA-NH2 forming first the succinimide on-pathway intermediate (Ac-Suc-GAA-NH2) thus, hydrolyzing to a product mixture of Ac-βDGAA-NH2 (isoAsp derivative) and Ac-αDGAA-NH2 (Asp derivative). Isomerization followed by 1H- (b) and 1H–15N-NMR (c) (700 MHz) as a function of the time. Both isoaspartyl (Ac-βDGRA-NH2) and aspartyl (Ac-αDGRA-NH2) as product mixtures, with the intermediate succinimide-derivative are observed by both 1D- and 2D-techniques beside the starting model system Ac-NGRA-NH2 (T = 55 °C and pH = 7.4). For TcN9 as for other proteins having longer backbone 2D/3D NMR is needed for resonance assignment and thus, for monitoring the isomerization rate. Resonances of TcN9 (blue) shift and self-duplicate giving a mixture of TcαD9 and TcβD9 (red and green) within a day. (d) The decay of 1H-NMR-signal intensities as a function of the time for selected resonances (e.g. acetyl protons: ~2 ppm), resulting in isomerization rate constant k 1, k 2, and k 3 at T = 310 K and pH = 7.8. (e) The same as (d) at T = 328 K and pH = 7.8.
Here we apply a nuclear magnetic resonance (NMR)-based approach using designed peptide-models within different molecular scaffolds that allow the in situ and real-time monitoring of the reaction. Deciphering key physicochemical parameters of the isomerization, we show how an appropriately rigid backbone fold can effectively delay such deleterious transformations.
Results and discussion
Effect of temperature and pH
To assess the effect of temperature (T = 28–55 °C) and pH (pH = 5.1–7.8) on the conversion rate, circular dichroism (CD) and NMR measurements were carried out focusing on both reaction kinetics and energetics. The reference peptide model, Ac-NGAA-NH2, has an isomerization half-life of about a day under physiological conditions (τ NGAA37 °C/pH=7.4 = 28.5 ± 3.9 h) (Supplementary Fig. 1, Fig. 1 and Tables 1 and 2). Lowering the pH increases τ exponentially, as exchange of the amide NHGly with hydrogens of water becomes slow: τ NGAA55 °C/pH=7.8 = 2.7 ± 0.5 h, τ NGAA55 °C/pH=7.4 = 4.1 ± 0.6 h, τ NGAA55 °C/pH=6.3 = 30.2 ± 2.9 h, and τ NGAA55 °C/pH=5.1 = 172.5 ± 14.4 h. Increasing T shortens τ exponentially (Table 2) shown in the case of the Ac-NGAA-NH2 and Ac-NGKA-NH2 model peptides. At a slightly elevated temperature deamidation-induced isomerization of a tetrapeptide is completed within a few hours: e.g. τ NGAA55 °C/pH=7.4 = 4.1 ± 0.6 h. The product ratio reflected in k 2/k 3 = 4.45 ± 0.3 shows that –βAsp– is more abundant than the –αAsp– product. The calculated activation energies (Table 2) also indicate that under physiological conditions the conversion reaction is indeed feasible. The determined energy differences are comparable to that reported for a peptide segment of the α A-crystalline (Aki et al., Reference Aki, Fujii and Fujii2013).
Table 1. Isomerization half-lives (τ) and related thermodynamic data for NGAA and NGRA model systems by employing the Eyring–Polányi equation

Pearson correlation coefficients of the linear fitting are: (a) 0.98, (b) 0.99, and (c) 0.95.
Table 2. Isomerization half-lives (τ) of Asn/Asp (at pH = 7.4) built in different and tunable molecular scaffolds

a τ E(–)NGK(+)37 °C/pH=7.4 ~ 32.0 ± 8.4 h.
b τ K(+)NGE(−)37 °C/pH=7.4 ~ 45.2 ± 8.1 h.
c τ WVVW37 °C/pH=7.4 = ~ 79.6 ± 10.6 h.
d τ NGAA55 °C/pH=5.1 = ~172.5 ± 14.4 h; τ NGAA55 °C/pH=6.3 = ~30.2 ± 2.9 h; τ NGAA55 °C/pH=7.8 = ~2.8 ± 0.5 h.
e τ NGRA55 °C/pH=5.1 = ~ 171.5 ± 15.7 h.
f τ TcN937 °C/pH=7.4 = ~ 40.0 ± 5.0 h.
g Experiment repeated three times.
h The 20-residue-long Trp-cage protein, TcD9 (Neidigh et al., Reference Neidigh, Fesinmeyer and Andersen2002).
i Values taken from these studies.
Changing the chemical constitution at the reaction center (i) and at neighboring positions ((i + 1) and (i + 2))
Negative charge significantly slows the isomerization rate: Asp at the i th-position isomerizes with a much slower rate than Asn within the same molecular framework: Ac-D(–)GAA-NH 2 isomerizes >33 times slower than Ac-NGAA-NH 2 does. Next neighbor effects on the deamidation rate (position (i + 1)) were studied extensively (Robinson and Robinson, Reference Robinson and Robinson2001a; Robinson et al., Reference Robinson, Robinson, Robinson, Robinson, Robinson, Robinson and Robinson2004). The rate of isomerization was shown to be faster if the residue following the Asn/Asp is small (e.g. Gly and Ser), slower for large and hydrophobic residues (e.g. Ile, Leu, Phe, Val, and Trp) and practically impossible for Pro (Robinson and Robinson, Reference Robinson and Robinson2001a; Robinson et al., Reference Robinson, Robinson, Robinson, Robinson, Robinson, Robinson and Robinson2004). In accordance with mass spectrometry data, our NMR results also demonstrated that if Gly(i+1) is replaced by bulkier residues (Ala, Ser, and His), τ lengthens significantly: compared to that of the Ac-NGAA-NH 2 reference system, in the His < Ser < Ala order (Supplementary Table 3). Considering the (i + 2) position, it was previously postulated that the His, Arg, and Lys residues at this site will increase the reaction rate considerably (Robinson and Robinson, Reference Robinson and Robinson2004c), as a proton donor can approach and stabilize the negative reaction center, [–(C=O)–N–](−) of the backbone amide and thus facilitate isomerization (Goolcharran et al., Reference Goolcharran, Stauffer, Cleland and Borchardt2000) (Supplementary Scheme 1). Our results confirm this hypothesis – we found that if a charged residue (Arg( + ) or Lys( + )) follows the –NG– motif at the (i + 2) position, τ shortens considerably. For Ac-NGR(+)A-NH 2 and Ac-NGK(+)A-NH 2 shortening is ~25 and ~32%, respectively. If the positive charge is placed closer to the reaction center, τ gets even shorter: cf. (H+)-NGAA-NH 2 (Supplementary Table 3), but the catalytic effect is diminished if it is distantly located at the (i + 3) position (cf. Ac-NGAR(+)-NH 2). This effect is even more enhanced when less potent (i + 1) residues are present (e.g. Ala and Ser), the well-placed positive charge speeds up the reaction in all cases significantly: τ is shortened by ~55% when comparing –NAR(+)A– to –NAAA or –NSR(+)A to –NSAA variants (Supplementary Table 3, Fig. 2). A second positive charge has a marginal effect (cf. (H + )-NGR(+)A-NH2). However, amphoteric His neither enhanced nor reduced isomerization rate (cf. Ac-NGHA-NH 2).

Fig. 2. Constitutional changes at i, (i + 1), and (i + 2) positions with respect to Asn significantly alters the isomerization rate. The introduction of a negative charge Asn→Asp(−), as well as an increase of the side-chain bulkiness at the (i + 1) position reduces the isomerization rate by magnitude(s). In contrast, a positively charged side-chain at (i + 2) enhances the rate of isomerization by a factor of ~2.
It is important to note that the above is only true in the case of Asn as the i th residue, the isomerization rate of Asp(−) is not enhanced by a positive charge at the (i + 2) position, (compare Ac-DGAA-NH 2 and Ac-DGRA-NH 2 in Supplementary Table 3), most likely due to salt-bridge formation between long, flexible side chains carrying a positive charge and either of the carboxylate moieties.
The effect of conformational mobility at the reaction center
Compared to Gly, both small apolar (Ala) and polar (Ser and His) side chains reduce the available backbone conformational space at (i)-(i + 2) positions significantly (Perczel et al., Reference Perczel, Angyan, Kajtar, Viviani, Rivail, Marcoccia and Csizmadia1991b; Perczel et al., Reference Perczel, Farkas and Csizmadia1996). Therefore, the above-observed reduction in the reaction rate can originate either from constitutional (different molecular scaffolds and polarities) and/or conformational (different internal mobility) effects. Ac-ANGA-NH 2 partly adopts a type I′ or I/II′ β-turn (Perczel et al., Reference Perczel, Hollosi, Foxman and Fasman1991a; Perczel et al., Reference Perczel, McAllister, Csaszar and Csizmadia1993; Hutchinson and Thornton, Reference Hutchinson and Thornton1994) unlike the fully unfolded Ac-NGAA-NH 2 and has a ~30% longer τ, signaling that increasing backbone rigidity slows the isomerization rate (Supplementary Table 3). The salt-bridge enforced β-turn fold of Ac-E(−)NGK(+)-NH 2 has a similarly longer τ as compared to Ac-NGAA-NH 2, while the swapped Ac-K(+)NGE(−)-NH 2 variant an even longer (~66% increase) (here, in addition to the restriction on backbone motions, the reaction enhancing positive charge of the Lys at the (i + 2) position is replaced by the negative center of Glu). Furthermore the conformational freedom of –NG– is reduced even more as within H-SWTVE(−)NGK(+)VTWK-NH 2, a Trp-paired zipped β-hairpin structure (Wu et al., Reference Wu, McElheny, Huang and Keiderling2009), we observe that the restricted internal dynamics increase the halftime of the isomerization over 2.5-fold (Supplementary Table 3). Similarly, placing the NGAA-motif of the highly flexible Ac-NGAA-NH 2 into that of cyclo(NGAANGAA) with limited internal dynamics and into the conformationally frozen cyclo(NGAA), resulted in τ increases of 2.5- and 5.3-fold, respectively (Supplementary Table 3 and Fig. 3). A significant decrease of conformational variability was confirmed in our molecular dynamics (MD) simulations where the Ac-NGAA-NH 2 samples 101, the cyclo-octapeptide 40, and the cyclo-tetrapeptide only three conformations along the equilibrium trajectory (Supplementary Fig. 1). Moving in the reverse direction, conformational restriction can be reduced by elevated T. Accordingly, the half-life of isomerization of the β-hairpin (H-SWTVE(−)NGK(+)VTWK-NH 2) decreases over 6-fold as the temperature is raised from 310 to 328 K, with a coupled decrease of fold compactness from 60 to 35% (Fig. 4a).

Fig. 3. As conformer heterogeneity of selected oligo- and polypeptides and protein models decreases (fewer clusters), the fold dependent protective factor increases and thus, isomerization rate gets reduced (τ increases). MD conformers were clustered and superimposed based on the backbone structure of the X-NG-Y motif. Mid-structures of the most populated clusters accounting for 90% of all the snapshots are shown in the case of the dynamically and conformationally ‘locked’ NG-subunits: (a) 19 for the ‘free’ Ac-NGAA-NH2, (b) 6 for the restricted cyclo(NGAANGAA) and (c) 1 for the ‘locked’ cyclo(NGAA). (d) 20 for the ‘free’ Ac-ENGK-NH2, while (e) 10 for the β-hairpin model of limited internal motion containing the –ENGK– motif. (f) 27 for the unrestrained Ac-KNGG-NH2, but only (g) 11 for the folded TcN9 mini-protein hosting the same –KNGG– but ‘locked’ motif. Asn and Gly are shown explicitly in cyan and green.

Fig. 4. Fold-compactness determines the magnitude of the protective factor. The well-folded β-hairpin (a) and the TcN9 protein (h) restrict the backbone motion around NG, compared to the unrestricted backbone of Ac-ENGK-NH2 (b) and Ac-KNGG-NH2 (i). Asn and Gly are shown explicitly in cyan and green, respectively. The folded (black)/unfolded (gray) ratio (%) of the conformational ensembles at different temperatures derived from their far UV-electronic circular-dichroism (ECD) spectra (Perczel et al., Reference Perczel, Hollosi, Tusnady and Fasman1991c; Perczel et al., Reference Perczel, Park and Fasman1992). Folded % shown as black bars (c, e, j, and l) with the reference pure ECD curves centered (d and k). Integral changes of selected 1H-NMR-resonances (e.g. HβAsn) as function of the time (f, g, m, and n) enable to calculate isomerization half-lives: τ(h). Isomerization half-lives (pH = 7.4) of the four systems at two temperatures (37 and 55 °C) show that as the folded content drops (60→35%) and (66→43%) at higher T(°C)s (c, e, j, and l), the relevant τ also decreases (79.6 ± 10.6→13.0 ± 0.6 h) and (40.0 ± 5.0→5.4 ± 0.6 h), demonstrating the facilitation of the isomerization.
We thus found that a rigid backbone can act as an effective protective factor, which leads to the conclusion that –NG– motifs that are preserved during evolution, especially in long-lived proteins, must have a higher than average structural rigidity. There are NMR descriptors, measuring rigidness of the protein backbone, the <S 2> and <hetNOE> values, available from the Biological Magnetic Resonance Bank (BMRB) (Ulrich et al., Reference Ulrich, Akutsu, Doreleijers, Harano, Ioannidis, Lin, Livny, Mading, Maziuk, Miller, Nakatani, Schulte, Tolmie, Kent Wenger, Yao and Markley2007). S 2 of an unstructured residue is ~0 and it is around 1 if the NH-vector nutation is fully restricted, while nuclear Overhauser effect (NOE) enhancement of the NH bond is around −3.9 for fast and 0.8 for slow motions. We found 207/225 S 2 and 224/289 hetNOE data values referring to AsnGly/AspGly units, with average values of <S 2>all.NGs = 0.87 ± 0.13 and <S 2>all.DGs = 0.84 ± 0.14, as well as <hetNOE>all.NGs = 0.67 ± 0.34 and <hetNOE>all.DGs = 0.73 ± 0.22. Although Gly typically introduces higher backbone mobility affecting neighboring residues as well, we found that almost all (>90%) NG/DG motifs in BMRB have rather rigid backbone structures. Only seven out of the above cases presents higher than average mobility for –NG–. Likewise, among the 145 analyzed cases of –DG– motifs, over 134 (>95%) are in a rigid backbone environment and only three show elevated internal mobility. Thus, if an NG/DG motif appears in a sequence, it is preserved as a rigid unit, in spite of the presence of the inherently flexible Gly residue. However, the stabilization effect of such rigidity is not always fully effective – exceptions can also be found. For example, the isomerization of the –NG–motif of a Tyr phosphatase protein (BMRB entry 19388 (Stehle et al., Reference Stehle, Sreeramulu, Lohr, Richter, Saxena, Jonker and Schwalbe2013) or 26513 (Stehle et al., Reference Stehle, Sreeramulu, Lohr, Richter, Saxena, Jonker and Schwalbe2015)), even though both the Asn and Gly show restricted backbone dynamics (S 2Asn161 or Asn164 = 0.41 and S 2Gly162 or Gly165 = 0.18), takes place, nevertheless (Supplementary Table 1).
Effect of the local conformation
Another interesting, apparent counter-example to the concept that a stable fold provides a safeguard against isomerization in our experiments is the case of transferring the –KNGG– subunit into the well-folded Trp-cage protein (TcN9: NLYIQWLK(+)NGGPSSGR(+)PPPS) (Neidigh et al., Reference Neidigh, Fesinmeyer and Andersen2002). In this case only a moderate increase of τ was found between the free and the protein entrapped form (Fig. 4b) even though the NG unit was placed at the end of the α-helical segment. The conformational rigidity of the NG unit in this matrix matches that of the previously mentioned β-hairpin model (with 64 (TcN9) and 60 (β-hairpin) conformations sampled during their 328 K MD simulations), but the protection factor of this specific fold is characteristically less pronounced (by a factor of 2, as compared to that of the β-hairpin). Thus it appears that beyond rigidity or flexibility, the specific arrangement of the reacting partners of the deamidation within a given fold should also be considered. The orientation and distance of the nucleophilic amide and its target Cγ = Oδ can be described using Bürgi–Dunitz (BD) distance (d) and angle (θ) to track the isomerization path (Burgi et al., Reference Burgi, Dunitz, Lehn and Wipff1974; Ospina and Villaveces, Reference Ospina and Villaveces1993) (Fig. 5). The rate-limiting step of the reaction is the succinimide ring-closure taking place in the [ψ,χ 1] conformational subspace of Asn. In the protein data bank (PDB), several structures can be found where such succinimides of isomerization-prone Asn residues were captured in the protein matrix. It is important to note that the θ and d values are very similar in these proteins, not only in their the succinimide-state [θ ~ 125o, d ~ 1.30 Å], but also in their unreacted form, where their BD parameters are scattered within a well-defined region of d = 3.25 ± 1.2 Å, θ = 109.5 ± 10° (Fig. 5a), regardless of the protein type (hen egg-white lysozyme, legumain, apo-CheY, endothiapepsin, amylomaltase), sequential motif or secondary structure that hosts the specific Asn. In fact, the Asn-Gly units that eventually transformed into a succinimide are embedded in very different local environments: such as in α-helix, β-pleated sheet, β-turn, loops, etc. (Figs 5c and d) suggesting that a rigid structure does not, in all cases, provide protection against deamidation – not if the conformation of the Asn-Gly units are located within the reactive zone of the BD diagram. It is also interesting to see that in those proteins where the NG-motif was reported to undergo deamidation but with a half-life that is long enough to allow purification and crystallization procedures (minimum of several weeks) – where succinimide intermediates were not detected –, the θ and d values were found outside the reactive zone of the BD diagram (Fig. 5b), while appearing in virtually all regions of the Ramachandran plots (Figs 5c and d). This also indicates that an NG-motif, accommodated in even quite unfavorable local and secondary structure settings will eventually isomerize – the difference is only a matter of time (Tompa, Reference Tompa2010).

Fig. 5. Variables d(Å), θ(°), χ 1(°), and ψ(°) of Asn/Asp are shown to describe ring-closure, the rate-limiting step of the isomerization. (a) Red dashed oval (d = 3.25 ± 1.2 Å, θ = 109.5 ± 10°) encircles PDB conformers for which isomerization is possible because of the spatial vicinity of the nucleophile NGly and the reaction center Cδ = Oγ. The red circle encompasses PDB conformers where succinimide intermediates were crystallized and their structures were determined, namely hen-egg-white lysozyme or HEWL (green), legumain (blue); apo-CheY (magenta); endothiapepsin (cyan); and amylomaltase (orange). (PDB codes are as follows: HEWL: 1gwd, 1gxv, 1gxx, 1h6m, 1h87, 1hf4, 1w6z, 2blx, 2bly, 2bpu, 2c8o, 2c8p, 2cgi, 2w1l, 2w1m, 2w1x, 2x0a, 2xbr, 2xbs, 2xjw, 2xth, 2ybh, 2ybi, 2ybj, 2ybl, 2ybm, 2ybn, 2ydg, 3zvq, 4a7d, 4aga, Legumain: 4aw9, 4awa, 4awb, 4fgu, 4nom, apo-CheY: 3rvk, 3rvq, Endothiapepsin: 1e5o, Amylomaltase: 1esw.) (b) A similar (d, θ)-plot as A for proteins of which isomerization was described, but succinimide intermediates were not yet isolated: –VNGP– of α B-crystallin (black) [2klr] and –GNGR– of GroES (light green) [1pcq] are both part of β-pleated sheets, –ENGA– of Ser-hydroxymethyl-transferase (green) [1rv3] is situated in an α-helix, –SNGP– of SOD1(red) [1rk7] as well as –ENGK– and –KNGE– of β 2-microglobulin (magenta and yellow) [1jnj] are integrated into β-turns and –ENGE– of GroeS (blue) [1pcq], –GNGY– of calbindin D28 (cyan) [2f33], –GNGR– of fibronectin (orange) [1fbr] are part of loops. (c) and (d) Asn associated (χ 1, ψ) and (ϕ, ψ) 2D-plots of the isomerizing NG-units of the above proteins: dot represents Asx-conformers, while triangles for the associated succinimide. Note that these examples cover most major secondary structural elements, namely the α-helix, β-pleated sheet, β-turn, and loops.
To better understand the geometrical criteria of Asn/Asp isomerization, we carried out rigid-body modeling of Ac–ANGA–NH 2 using the optimum value of θ ( = 109.5 ± 1°). The topology of the obtained potential energy surface reveals that for a very wide range of χ 1 and ψ values, optimal reactive condition (d < 3.25 Å and θ = 109.5 ± 1°) can be reached (Fig. 6(I)): here succinimide formation would be prompt with short τ. The accessibility of such arrangements is well reflected by the fact that analyzing all protein families represented in the PDB (Berman et al., Reference Berman, Westbrook, Feng, Gilliland, Bhat, Weissig, Shindyalov and Bourne2000) (using a homology filtered set of 2724 entries), 42.6% of a total of 57925 AsnXxx/AspXxx-motifs fall in the reactive zone (d < 4.25 Å) and 9.4% within the highly reactive zone (d < 3.5 Å) (Fig. 6(II)). The very center of this zone holds the succinimide derivatives that were isolated and crystallized already (Fig. 6(II)). Still within the 3.25 < d < 4.25 Å zone special Asx-turns can be assigned: categorized as type I, I′, II, and II′ Asx-turns, each analogous to the appropriate β-turn (Duddy et al., Reference Duddy, Nissink, Allen and Milner-White2004). They act as ‘gatekeepers’ of isomerization as they have relatively short d values, (3.25 < d < 4 Å), and while their θ values are unsuitable for isomerization (typically: 0 < θ < 90° or 130 < θ < 180°), the inherent flexibility arising from NH X-s that do not participate in H-bond networks, allows both χ 1 and χ 2 to adopt a conformation promoting isomerization. For 57.4% of all AsnXxx/AspXxx-motifs of the homology filtered protein structures, d is greater than 4.25 Å, so in these cases, even if θ is optimal, succinimide formation would be slow. Common secondary structures of this region are γ inv-turn (or γ L), δ L- and left-handed α-helix (or α D) (Perczel et al., Reference Perczel, Hollosi, Foxman and Fasman1991a), so these are conformations that could delay isomerization considerably. On the other hand, analysis of the Ramachandran-surface, f(&phis;, ψ) (Fig. 6/III) of all Asn/Asp residues of the homology filtered set shows that both the β-valley (β-strands, polyPro II, etc.) and the α-helical regions contain zones colored red for which d < 3.25 Å and θ = 109.5 ± 1°, and thus, both succinimide formation and the subsequent isomerization would readily proceed (even if the NHGly needed for the nucleophilic attack is engaged in the fold stabilizing H-bond pattern). Residues populating the surrounding blue/gray zones of the Ramachandran-surface are expected to isomerize more slowly (longer d- and/or perhaps less optimum θ-values). This analysis clearly demonstrates that even NX/DX-motifs that are ‘locked’ into secondary structural elements might adopt conformations optimal for isomerization.

Fig. 6. (I) Distances (NGly to CGAsn) at the optimal BD angle (NGly-CGAsn-OD1Asn) as a function of the χ 1Asn and ψ Asn torsions, of the rigid-body model of Ac-ANGA-NH2 shows a single valley with centered at f(120°; 240°). Red-magenta (d < 3.25 Å) and blue-white regions (3.25 Å < d < 4.25 Å) outlined with green lines indicate distances within the ‘reactive zone’. The darker the gray (d > 4.25Å) the more unreactive the structures are. Note that a reactive distance can be reached at a very wide range of ψ Asn. (II) The f(χ 1Asx,ψ Asx) surface of –AsxXxx– selected from non-homologous proteins of the PDB with the same color-coding and region boundaries. Succinimides are shown as black triangles in the middle of the reactive valley. (III) Identical dataset mapped as d ~ f(ϕ Asn;ψ Asn) function, similar to a Ramachandran surface. Black triangles again represent succinimides found in the PDB. Distances and angles of the MD simulated equilibrium ensembles of (a) Ac-NGAA-NH2, (b) cyclo(NGAANGAA), (c) cyclo(NGAA), (d) Ac-ENGK-NH2, (e) β-hairpin model hosting the –ENGK– motif, (f) Ac-KNGG-NH2, and (g) the mini-protein TcN9 hosting the –KNGG– motif. Orange rectangles show the boundaries of the reactive zone, orange dots the median of the distributions.
Based on the above, it seems that beyond sequence and local flexibility/rigidity, the BD-parameters of the NX/DX-motif are crucial in determining the isomerization rate. Using these cornerstones, we could rationalize the NMR derived half-life of our model systems based on their MD derived conformational heterogeneity (backbone flexibility of the four-residue-stretch hosting the NG-motif) and BD-parameter distribution (Figs 3 and 6). The highly flexible Ac-NGAA-NH 2, Ac-ENGK-NH 2, and Ac-KNGG-NH 2 tetrapeptides sample 101, 95, and 122 different backbone conformations respectively. Although the median values of θ and d (~92° and 3.7 Å) of these systems fall outside the reactive zone of the BD diagram (θ = 109.5 ± 10°, d = 3.25 ± 1.2 Å), its inner territory is well represented in the equilibrium ensemble as well. As discussed earlier, while the NGAA-motif within a cylo-octapeptide and a cyclo-tetrapeptide scaffold becomes increasingly rigid, its BD-descriptor values shift toward less favorable regions: θ/d medians are 91°/3.70 Å and 81°/3.80 Å, respectively. Especially in the case of the cyclo-tetrapeptide scaffold, the combination of a rigid backbone fold with unfavorable BD-parameters gives the perfect example of an off-pathway structure of the isomerization pathway, isomerizing >5 times slower than the unbound free form. Similarly, placing the ENGK-motif into a β-hairpin scaffold the backbone conformation becomes more rigid with BD-values (θ/d medians are ~94°/4.1 Å) representing an off-pathway structure and thus, isomerization gets slower with τ increased by ~2.6 fold. On the other hand, building the KNGG-motif into TcN9, at the tip of the α-helical segment of the Trp-cage fold, causes a significantly lower increase in τ (1.3 fold) in spite of the similar extent in the reduction of backbone mobility. However, in this molecular scaffold, the median of the BD-descriptors (θ/d ~ 102°/4.20 Å) is clustered near the reactive zone, the Trp-cage fold thus provides an example of an on-pathway fold, and where the restricted backbone fixes the reaction partners into a geometry that promotes the isomerization of Asn.
Pathophysiological significance
Deamidation-induced isomerization of Asn/Asp residues has been called a molecular clock (Robinson and Robinson, Reference Robinson and Robinson2001a), ticking as a time-bomb threatening proteins' structural integrity. The conversion of Asn is irreversible as NH3 is released (and lost) during the process, while that of Asp can and is reverted (Clarke, Reference Clarke2003) in cells by protein l-isoaspartyl/d-aspartyl-O-methyltransferase (PIMT) (O'Connor, Reference O'Connor2006), an evolutionarily highly conserved enzyme typically repairing βAsp modifications. The severity of –NG– and –DG– isomerization associated malfunctioning is underlined by the ubiquitous expression of PIMT/PCMT in both pro- and eukaryotic cells. The presence of PIMT in a wide range of taxa highlights its functional importance (Skinner et al., Reference Skinner, Puvathingal, Walter and Friedman2000) showing that evolutionary tolerated Asx-isomerization sites will in all cases be guarded – most effectively in highly flexible segments where the spontaneous isomerization is also the most straightforward.
When a protein's half-life is shorter than the isomerization half-time of its AsnXxx/AspXxx units (τ isomer > τ protein), the protein is degraded long before any isomerization might complete. Human ornithine decarboxylase [UniProt ID: P11926] is such an example (τ protein ~ 11 min) containing two AsnGly units (N126 G and N398G), which both might isomerize rapidly due to their favorable BD values (θ N126G ~ 120° and θ N398G ~ 136°) but without being a risk to the proteome. But the opposite holds if τ isomer < τ protein, presenting a considerable challenge in maintaining homeostasis (Supplementary Table 2). For example, human hemoglobin α and β subunits of the erythrocytes circulate for ~100–120 days in adults (Harrison, Reference Harrison1979) and contain four and five Asn residues per subunit, respectively. In the most frequent alleles, there is no AsnGly unit found: instead of Gly, Val(N10V), Leu(N81L), Phe(N98F), and Ala(N79A) residues are located after Asn (Weintraub and Deverman, Reference Weintraub and Deverman2007). Furthermore, as all these AsnXxx motifs are part of α-helices, the implemented structural restrain further reduces the isomerization rate. However, several SNPs (single nucleotide polymorphisms) result in sensitive hemoglobin mutants, among which the Sardegna (N50H→N50G), Singapore (N78A79→D78G79), Wayne (K139Y→N139T), La Roche-sur-Yon mutation (N80L→N80H), and Providence N or D (K82G→N/D82G) present extreme vulnerability, enhancing the isomerization rate and causing detectable changes in flexibility near the heme binding site (Wajcman et al., Reference Wajcman, Kister, Vasseur, Blouquit, Trastour, Cottenceau and Galacteros1992) and are all associated with inherent blood disorders (e.g. hemolytic disease). The crystal structure of a Providence (K83G→D83G) mutant was determined (PDB: 5sw7) and shows the local conformation quite favoring isomerization (θ D83 ~ 109° and d ~ 4.6 Å) with NHGly free to exchange with water, located at the edge of an α-helix with a relatively dynamic backbone.
As for crystallins of the eye-lens, no turnover is possible (τ protein ~ 80 years) therefore N78L- and N146G-sites present a considerable vulnerability. Due to the large and hydrophobic side-chain of Leu, N78L is expected to isomerize slowly: a reaction further hindered by the low-backbone mobility of its location in a β-strand with its backbone amide NHs strictly cross H-bonded. However, N146G poses a more real threat (Supplementary Table 3). Even though its θ is outside the optimum range (θ N146 ~ 80°), its backbone amide NH is H-bonded and thus becomes a poor nucleophile and it is located in a β-strand in which little room is left for backbone dynamics, the Asn→βAsp conversion does take place and leads to the loss of the transparency and refractivity of eye-lens and cornea (i.e. age-related cataract) through promoting aggregation (Fujii et al., Reference Fujii, Takata, Fujii and Aki2016). Proteopathies linked to hemoglobin, crystallin, etc. lead to the concept of chronoregulation (Weintraub and Deverman, Reference Weintraub and Deverman2007), which seems to be essential for life.
On the other hand, isomerization of Asn to Asp may also be part of regulatory functions. The anti-apoptotic Bcl-xl contains a PEST degradation signal: TEAPEGTESEMETPSAIN52GNPSW that becomes considerably more potent by the N52G→D52 G isomerization (of τ isomer ~ 20 h) (Rooswinkel et al., Reference Rooswinkel, van de Kooij, de Vries, Paauwe, Braster, Verheij and Borst2014). The change leads to a more effective ubiquitin-directed degradation (Rechsteiner and Rogers, Reference Rechsteiner and Rogers1996) of the protein, which in turn initiates cell death (Zhao et al., Reference Zhao, Oxley, Smith, Follows, Green and Alexander2007; Dho et al., Reference Dho, Deverman, Lapid, Manson, Gan, Riehm, Aurora, Kwon and Weintraub2013): thus the presence of the NG-motif creates an active mode of age surveillance. Another example of regulation, in this case through cellular targeting, is the flexible and conserved GNG N-terminus of PKAs that will no longer be myristoylated if the conversion to GDG takes place (Jedrzejewski et al., Reference Jedrzejewski, Girod, Tholey, König, Thullner, Kinzel and Bossemeyer1998).
Concluding remarks
Here we have shown that the decreased mobility of compact and rigid backbones of proteins acts as a protective factor against spontaneous deamidation and isomerization at their Asn/Asp-Gly ‘Achilles' heels’ and thus, helps to preserve their molecular integrity. However, our results also indicate that even within well-defined secondary structure elements, optimal conditions for isomerization can take place. Optimal BD-parameters and thus on-pathway structures can be found within turns, helices, and sheets. Several protocols were published to estimate the relative stability of an Asn in a given sequential (Robinson and Robinson, Reference Robinson and Robinson2004c; Lorenzo et al., Reference Lorenzo, Alonso and Sánchez2015) or structural (Robinson and Robinson, Reference Robinson and Robinson2001b; Lorenzo et al., Reference Lorenzo, Alonso and Sánchez2015; Jia and Sun, Reference Jia and Sun2017) environment that consider the chemical nature of the (i + 1) residue, the length of d, or whether the Asn is part of an α-helix/β-sheet. Here we described that it is the electrostatic contribution of the (i-1) and especially (i + 2) positions along the d(Å) and θ(°) BD-values and ultimately the flexibility of the molecular scaffold that are the key factors in determining the rate of this spontaneous isomerization. Our results indicate that all these should be considered when assessing the severity of mutations or designing protein drugs, antibodies and determining their optimal storage conditions (with respect to isomerization rate, it is advised to store proteins at moderately acidic media (subcutan pH ~5), unless this pH is close to the isoelectric point of the protein (where unwanted precipitation can destroy 3D-fold and reduce solubility)).
Materials and methods
Synthesis of linear and cyclic peptides
Linear NGR peptides were prepared on Rink-Amide 4-(2′,4′-dimethoxyphenyl-Fmoc-aminmethyl)-phenoxyacetamido-methylbenzhydryl amine resin resin (0.67 mmol g−1 capacity), while precursors for cyclization on 2-chlorotrityl chloride resin (0.6 mmol g−1 capacity) using the Fmoc/tBu-strategy, but the Fmoc-group was detached under milder conditions: 2% 1,8-diazabicyclo[5.4.0]undec-7ene/2% piperidine/0.1 M hydroxybenzotriazole (HOBt) in dimethyl-formamide (DMF) (6 times) to avoid succinimide ring formation. Semi-protected peptide was removed from the 2-ClTrt resin with the cleavage mixture (AcOH–MeOH–dichloromethane = 1:1:8 (v/v/v)), stirred for 2 h, at RT, filtered and evaporated. Products were purified by reverse phase high performance liquid chromatography (RP-HPLC): semi-preparative Phenomenex Luna C18 column with eluent A (0.1% trifluoroacetic-acid (TFA)/H2O) and B (0.1% TFA in MeCN/H2O (80/20)) and identified by ESI-MS (Bruker Esquire 3000+ ion-trap).
The preparation of head-to-tail cyclopeptides was usually performed by incorporating Gly to the C-terminus of linear precursor peptide to avoid racemization cyclization. However, for c[NGAA], precursor H-AN(Trt)GA-OH instead of H-AAN(Trt)G-OH was used to get the cyclic tetrapeptide. Cyclization was performed in DMF in the presence of benzotriazol-1-yloxytris(dimethylamino)phosphonium hexafluorophosphate/HOBt/N,N-diisopropylethylamine (12/12/24 eq.) overnight at 40 °C. The reaction was stopped (with TFA/DMF), and purified by RP-HPLC and characterized by electrospray ionization mass spectrometry. The main product (~90%) was c[N(Trt)GAAN(Trt)GAA] with <10% of c[N(Trt)GAA]. Side-chain deprotection achieved at room temperature with 95% TFA/2.5% TIS/2.5% H2O for 1.5 h. Once precipitated with Et2O, the product was purified with RP-HPLC.
Protein expression labeling and purification
Tc9N's complementary deoxynucleic acids (cDNAs) were ligated into a SacII and BamHI site of the pUBK2 vector, and the plasmid encoding H10-Ub-Protein was transformed into the Escherichia. coli strain BL21(DE3) (Stráner et al., Reference Stráner, Taricska, Szabó, Tóth and Perczel2016). Transformed cells were grown in Luria-Bertani medium containing 0.1 mg ml−1 Kanamycin (200 rpm; 37 °C). Cells (OD600 0.6–0.8) were isopropyl β-D-1-thiogalactopyranoside induced to a final concentration of 1 mM and harvested after 3 h incubation (37 °C), resuspended and lysed in buffer (300 mM NaCl, 50 mM NaH2PO4, and 3 mM NaN3) by sonication. Proteins were purified using a 5 ml nickel-nitrilotriacetic acid (Ni-NTA) chromatography column. The eluted fractions with fusion proteins were overnight dialyzed into a buffer and were 3–4 h digested with His-tagged yeast ubiquitin hydrolase (YUH). The hydrolyzates having monitored on sodium dodecyl sulfate–polyacrylamide gel electrophoresis, the ubiquitin and YUH were separated from the protein by Ni-NTA chromatography, purified by reverse-phase HPLC (C18 column), and identified by a Perkin–Elmer Sciex API2000 mass spectrometer.
BMRB analysis
hetNOE and S 2 parameter were analyzed for available proteins retrieved from the BMRB (Ulrich et al., Reference Ulrich, Akutsu, Doreleijers, Harano, Ioannidis, Lin, Livny, Mading, Maziuk, Miller, Nakatani, Schulte, Tolmie, Kent Wenger, Yao and Markley2007). From the 11817 proteins deposited, 772 have either T 1, T 2, hetNOE or S 2 values. In total, hetNOE for 289 proteins, while S 2 for 92 proteins were reported in the BMRB. Among these, 74/84 and 46/58 proteins have in total 115/145 and 76/151 –NG/DG– subunits of known hetNOE data and with S 2 parameters reported, respectively, once redundancy removed.
NMR experiments and assignment
For shorter peptides, 1H-NMR data and two-dimensional (2D) homonuclear (correlation spectroscopy, total correlation spectroscopy, and rotating frame nuclear Overhauser effect spectroscopy), for proteins 15N–1H- and/or 13C–1H-heteronuclear single-quantum correlation (HSQC) and heteronuclear (13C–1H-HSQC and heteronuclear multiple-bond correlation) spectra were recorded to analyze the requested kinetic information and to get both assignment and structural information, respectively. Time-dependent 1H-NMR measurements (Δt = 5, 10, 15, 20, 25, 30, 45, 60, 90, 120 min, etc.) were obtained to follow the isomerization (Supplementary Table 3) until the reactant concentration typically dropped below 10–20%. Isomerization (c ~ 3–5 mM) was followed by 1H-NMR (Bruker Avance 700 MHz): 9/1:H2O/D2O, 5 µl DSS and NaN3 (2 mg NaN3 and 1 mg DSS dissolved in 5 ml H2O), buffers: pH = 7.40 and pH = 7.80 (50 mM Na2HPO4 buffer), pH = 6.33 (50 mM Na2HPO4 and 50 mM NaH2PO4), and pH = 5.10 (20 mM CH3COONH4). T = 310, 319, and 328 K were applied (Supplementary Table 4). To identify both the α- and β-isoforms of Ac-NGAA-NH 2 and of Ac-NGRA-NH 2CoCl 26H 2O was added as a shift reagent [Co(II)]/[Asp] ~ 0.023 (Supplementary Table 5) (Pivcová et al., Reference Pivcová, Saudek and Drobnik1982).
NMR data based kinetic simulation and thermodynamics
NMR signals were referenced to DSS. Selected 1H-NMR resonances (typically 8 to 12 per molecules) were simultaneously analyzed and the rate coefficients of the rate-limiting step, k 1, that of the succinimide formation, was determined, also expressed in terms of half-lives, or τ.
Succinimide hydrolysis leading to –βAspGly– and –αAspGly– have rate coefficients k 2 and k 3, respectively. The ratio of k 2/k 3 shows also the relative ratio of β- and α-isomers. Using the Eyring–Polányi equation, ln(k 1) plotted against T −1. The slope of the linear gives the enthalpy change, whereas the ordinate intercept provides the entropy change of the first step.
CD spectroscopy
Cd spectra of the proteins (35–150 µmol) were recorded on a Jasco-J810 spectrophotometer (1.0 or 10 mm cuvettes). Typical spectral accumulation parameters were at a scan rate of 50 nm min−1 with a 1 nm bandwidth and a 0.2 nm step resolution over 185–260 nm (far-ultraviolet (UV)) and 250–320 nm (near-UV) with four scans averaged for each spectrum at temperatures from 5 to 85 °C with 5 min/5 °C thermal equilibration. The solvent spectra were used as reference baselines and subtracted from the spectra. Conversion into mean residue molar ellipticity units ([Θ]MR, deg × cm2 × dmol−1) were conducted for the far-UV region. Quantitative spectral deconvolution was achieved by CCA+ software (Perczel et al., Reference Perczel, Hollosi, Tusnady and Fasman1991c).
Molecular dynamics simulations
MD simulation was carried out as implemented in GROMACS59, using the AMBER-ff99SBildnp* force field. The system was solvated with TIP3P water molecules in dodecahedral boxes with a size allowing 10 Å between any protein atom and the box. Total charge was neutralized and physiological salt concentration was met (Na+, Cl−). Energy minimization of starting structures was followed by sequential relaxation of constraints on protein atoms in three steps and an additional NVT step (100 ps) to stabilize pressure. Trajectories of 600–1200 ns NPT simulations at 310 and 328 K and 1 bar were recorded for further analysis (collecting snapshots at every 4 ps) (for backbone RMSD values see Supplementary Fig. 1). The last 200–600 ns of the trajectories was clustered fitting the main chain atoms using a 1 Å cutoff.
Rigid body modeling
Protected ADGA tetrapeptide was optimized at the B3LYP/6-311++G(d,p) level of theory. Rotations of all four dihedral angles (φ, ψ, χ 1, and χ 2) of the optimized Asp by 5 degrees and all NGly-CγAsp distances (d) and BD angles (θ) were determined. The structures having filtered on near-optimal 109.5° BD angles, ψ, χ 1 pairs, and NGly–CγAsp distances were plotted.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S003358351900009X.
Acknowledgement
The authors thank Viktor Farkas and Berill Ónodi for the WVVW hairpin and Pál Stráner for the mini protein model, Masashi Yokochi for the BMRB data, and Ernő Keszei and László Nyitray for scientific discussion. NMR spectrometer measurement time (700 MHz Bruker) was the courtesy of MedInProt Grant Facilitating Access to Instruments from the Hungarian Academy of Sciences. Calculations were carried out at the NIIF Supercomputing Center of KIFU (Hungary). This research project was supported by the European Union and the State of Hungary and co-financed by the European Regional Development Fund (VEKOP-2.3.3-15-2016-00009 and VEKOP-2.3.2-16-2017-00014), and the K116305 OTKA grant of the NKFIH of the Hungarian Academy of Sciences.