INTRODUCTION
Dairy cattle serve as an important reservoir for Salmonella and have been implicated in cases of human salmonellosis [1, Reference Zansky2]. The United States National Animal Health Monitoring System's Dairy ’96 study reported 5·4% of milk cows shed Salmonella and 27·5% of dairy operations had at least one cow shedding Salmonella [Reference Wells3]. Salmonella has been isolated from all ages of dairy cattle and throughout the production process. Mature dairy cattle typically appear asymptomatic while shedding this pathogen in their faeces [Reference Richardson4–Reference Edrington7] and while young calves are more susceptible to salmonellosis, cases in adult cattle have been reported [Reference Gay and Hunsaker8–Reference Anderson10].
Previous research conducted by our laboratory demonstrated significant variation in the prevalence of faecal Salmonella in healthy, lactating dairy cattle, not only among farms across the United States [Reference Callaway11], but also in farms within a small geographic area and in individual farms from season to season [Reference Edrington7]. Additional research examined production parameters (heifers vs. mature cows, lactation status, stage of lactation and heat stress) on Salmonella prevalence [Reference Edrington6, Reference Fitzgerald12]. While minor differences were noted in Salmonella shedding, results were generally inconsistent with no significant trends noted. Although heat stress did not result in any Salmonella shedding differences as measured by faecal incidence in the morning compared to the afternoon, in one experiment Salmonella prevalence averaged nearly 100% [Reference Edrington6].
Outbreaks of salmonellosis in mature lactating cows have been reported on some of the farms we sampled previously, resulting in substantial financial losses due to decreased milk production and cow mortality (personal communication with producers). Interestingly, these outbreaks are seasonal, occurring in late summer/early autumn, but are not always an annual event. One farm in particular had an increased incidence of Salmonella shedding, even in the winter when the incidence of Salmonella typically decreases [Reference Edrington7], compared to other regional dairies. In an attempt to elucidate factors involved in this seasonal salmonellosis, we examined Salmonella isolates obtained during an outbreak in 2003 from healthy and diarrhoeic cattle on this farm and then monitored 30 lactating cows, water, feed and pen soil samples on a monthly basis for 9 months the following year.
MATERIALS AND METHODS
2003 Salmonellosis outbreak
Faecal samples were collected during an outbreak of salmonellosis in the late summer of 2003 from mature lactating Holstein dairy cattle on a southwestern dairy farm (~2000 head). Cattle were maintained in large drylot pens, fed and managed as typical for dairy farms in this region of the United States. We were alerted to the outbreak in its final stages, therefore rectal grab samples could only be obtained from 10 diarrhoeic cattle that appeared to be in the early stages of salmonellosis. Ten apparently healthy lactating cattle from the same production group were also sampled at this time. We have sampled this dairy on multiple occasions [Reference Edrington7, Reference Fitzgerald12] and found Salmonella prevalence was often higher than neighbouring dairies and also quite variable from season to season in terms of overall prevalence, and serotype diversity. Based on our knowledge regarding Salmonella prevalence on this farm, we felt that sampling 10 healthy animals would provide for a reasonable assessment of the healthy cattle. If required, more samples would have been collected from healthy cattle the following week. All samples were cultured for Salmonella as described below.
2004 monthly surveillance
Mature multiparous Holstein dairy cattle on the farm described above were sampled on a monthly basis over a 9-month period (February to October 2004). Thirty head were selected that all calved within a 14-day period to eliminate potential animal differences associated with stage of lactation. Three cows were culled during the first 3 months of the experimental period from the sample population for various reasons, leaving 27 head remaining in the study group by October. Each monthly sampling was conducted in the morning with animals restrained in self-locking head stanchions. A palpation sleeve was utilized to obtain about 30 g faeces via rectal retrieval. Additionally, monthly samples of pen soil, water and the total mixed ration (TMR) were collected representative of the pens housing the study population of cows. Pen soil samples were collected from several locations within each pen using a soil probe which was disinfected between samples, from areas of full sun exposure and partial shade. Water samples were collected from each trough within a pen using sterile conical tubes. Multiple TMR samples (~100 g) were collected soon after feed presentation from the feed apron. Following each collection, all samples were placed on ice and shipped overnight to the USDA-ARS laboratory in College Station, TX for Salmonella culture and isolation described below.
Salmonella culture, isolation, serotyping and serogrouping
Salmonella was cultured by enriching about 10 g faeces in 90 ml tetrathionate broth (37°C, 24 h), followed by a second enrichment in Rapport–Vassiliadis broth (100 μl in 5 ml, 42°C, 24 h) prior to plating on Brilliant Green agar (BGA; Oxoid Ltd, Basingstoke, Hampshire, UK) supplemented with novobiocin (25 μg/ml). Following incubation (37°C, 24 h), five colonies exhibiting typical Salmonella morphology were randomly selected from each sample and confirmed biochemically using lysine and triple sugar iron agars. Positive samples were re-streaked on tryptic soy agar with 5% sheep blood (Becton, Dickinson and Company, Sparks, MD, USA) for further confirmation and serogrouping, using slide agglutination with Salmonella O antiserum (Difco Laboratories, Detroit, MI, USA). Salmonella isolates were stored (−80°C) using CryoCare™ bacterial preservers (Key Scientific Products, Round Rock, TX, USA). A portion of the isolates were sent to the National Veterinary Services Laboratory in Ames, IA for confirmatory serotyping. All media and agar were from Difco Laboratories (Detroit). Reagents and antibiotics were obtained from Sigma Chemical Co. (St Louis, MO, USA).
Pulsed field gel electrophoresis (PFGE)
Select Salmonella isolates were analysed by PFGE as described previously [Reference Hume13]. Briefly, Salmonella isolates were thawed and spread on BGA plates and incubated as described above. A single colony from each plate was selected and incubated overnight in 10 ml TSB, centrifuged (8000 g) and resuspended in 3 ml PBS. Washed cells were placed in a water bath (54°C) and mixed with equal volumes of 1·8% (w/v) low melting temperature agarose in PBS. Cells with agarose were transferred to disposable plug moulds for polymerization (4°C). Plugs were incubated overnight (50°C) in 20 ml lysis buffer [2% (w/v) sodium lauryl sarcosine; 1·0 m EDTA (pH 9–9·3); and 0·2 mg/ml proteinase K] before washing twice (20 min, 4°C) in TE [10 mm Tris (pH 8·0); 1 mm EDTA]. Plugs containing lysed cells were washed (3×; 20 min each) in 40 ml cold TE containing 40 μl phenylmethylsulfonyl fluoride (100 mm in isopropanol) and then washed three additional times (20 min) in cold TE. One half of each plug was incubated (20 min) with XbaI restriction endonuclease. Conditions for PFGE were: initial switch time=0·1 s; final switch time=90 s; included angle=120°; 6 V/cm; buffer temperature=12°C; run time=22 h. Genotypic relatedness was determined with Molecular Analysis Fingerprinting software, version 1.6 (Bio-Rad Laboratories, Hercules, CA, USA).
Antimicrobial susceptibility determination
Eighteen serotyped isolates each from the sick and healthy groups of cattle sampled in 2003 were examined for antimicrobial susceptibility using the Sensititre™ automated antimicrobial susceptibility system according to the manufacturer's directions (Trek Diagnostic Systems, Westlake, OH, USA). Broth microdilution was used according to methods described by the National Committee for Clinical Laboratory Standards (NCCLS) [14] to determine minimum inhibitory concentrations for the following antimicrobials: ampicillin, apramycin, ceftiofur, chlorotetracycline, enrofloxacin, florfenicol, gentamicin, neomycin, oxytetracycline, spectinomycin, sulphachloropyridazine, sulphadimethoxine, sulphathiazole, and trimethoprim/sulfamethoxazole. Resistance breakpoints were determined using the NCCLS interpretive standards [14] unless unavailable, in which case breakpoints in the United States National Antimicrobial Resistance Monitoring System (NARMS) 2000 Annual Report [15] were used. Escherichia coli ATCC 25922, E. coli ATCC 35218, and Enterococcus faecalis ATCC 29212 were used as quality control organisms.
RESULTS
A comparison of Salmonella isolates cultured from the faeces of diarrhoeic and healthy cows in 2003 is presented in Table 1. All 10 faecal samples from each group were positive for Salmonella and yielded 50 and 48 total isolates for the healthy and diarrhoeic cattle, respectively. Three of five serogroups (B, C1 and K) accounted for 84% of the isolates in the healthy cattle compared to the diarrhoeic cattle in which six of eight serogroups (C1, C2, E4, G, I, and K) represented 94% of the isolates. Serogroup C2 was not detected in any of the healthy Salmonella isolates while group B, which accounted for 22% of the healthy isolates, was not cultured in the faeceal samples of any of the sick cattle. Seven and nine different serotypes were identified in the healthy and sick isolates, respectively. The serotypes Senftenberg and Kentucky were not detected in any of the healthy cattle and accounted for 34% of the sick isolates. No differences in antimicrobial susceptibility patterns were observed in any the Salmonella isolates from sick and healthy cattle. Isolates were susceptible to all antimicrobials examined with the exception of spectinomycin, with three and five isolates resistant in the healthy and diarrhoeic groups, respectively (data not shown).
Table 1. Comparison of Salmonella isolates cultured from faecal samples collected from healthy and diarrhoeic lactating dairy cattle (2003)

PFGE was used to compare the genetic relatedness of isolates cultured from the faecal samples of healthy and sick cattle. Seventeen serotypes representing 84 isolates were examined. No genotypic differences were noted when comparing sick vs. healthy isolates (Table 2). Multiple genotypes within serotype were observed for a number of the isolates examined.
Table 2. Genotypes of selected Salmonella isolates cultured from healthy and diarrhoeic lactating dairy cattle (2003)

Monthly Salmonella prevalence in faecal, soil, TMR and water samples collected in 2004 are presented in the Figure. Diarrhoea was not observed in any of the 30 cows during any of the monthly sample collections. Faecal prevalence ranged from a high of 96% in August to a low of 19% in October, averaging 54% over the 9-month period. Pen soil and TMR samples were also highly variable in Salmonella prevalence ranging from almost zero to 100% positive. The percentage of soil and TMR samples positive for Salmonella over the 9-month sampling period averaged 39% and 76%, respectively. A high percentage of TMR samples positive for Salmonella did not always correlate to a high percentage of positive faecal samples. Water sampled from the pen troughs also varied, ranging from 0% to 75% positive, averaging 38% positive across the 9-month experimental period.

Fig. Salmonella prevalence (% positive) in faecal (■), soil (□), total mixed ration (![]() ), and water trough (
), and water trough (![]() ) samples collected monthly over a 9-month surveillance period at a southwestern dairy.
) samples collected monthly over a 9-month surveillance period at a southwestern dairy.
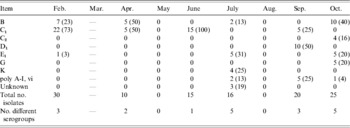
Faecal Salmonella serogroup prevalence is presented by month in Table 3. Groups B and C1 were most commonly identified, accounting for over 50% of the isolates each month except in August and October. In August, serogroup E4 replaced B as the predominant group, while in October E1, D1 and K accounted for 80% of the isolates. The number of different serogroups identified in the faecal samples ranged from a low of four in May to a high of 11 in March and August. Serogroups of TMR isolates were predominantly C1, E1 and E4 groups (Table 4). Multiple serogroups (four or more) were identified in TMR samples every month with the exception of July. The number of serogroups identified from soil isolates was low in the first 6 months of the study compared to the final 3 months (August, September and October) averaging 2·2 and 7·7 different serogroups, respectively (Table 5). Groups C1, B and K accounted for the majority of those identified from soil isolates over the 9-month period (Table 5). Similar to the other sample types, serogroup prevalence and diversity in water isolates varied by month over the course of the collection period, although in general fewer serogroups were identified (Table 6).
Table 3. Serogroup distribution [number and percentage of each month's isolates belonging to each serogroup (in parentheses)] of Salmonella isolates cultured monthly from faecal samples of healthy lactating dairy cattle over a 9-month period (2004)

Table 4. Serogroup distribution [number and percentage of each month's isolates belonging to each serogroup (in parentheses)] of Salmonella isolates cultured monthly from the total mixed ration (TMR) fed to healthy lactating dairy cattle over a 9-month period (2004)

Table 5. Serogroup distribution [number and percentage of each month's isolates belonging to each serogroup (in parentheses)] of Salmonella isolates cultured monthly from soil samples collected from pens housing healthy lactating dairy cattle over a 9-month period (2004)

Table 6. Serogroup distribution [number and percentage of each month's isolates belonging to each serogroup (in parentheses)] of Salmonella isolates cultured monthly from water samples collected from pens housing healthy lactating dairy cattle over a 9-month period (2004)
To examine serotype variation within an animal, we selected five cows that tested Salmonella positive (faeces) on the majority of the monthly samplings and randomly selected five isolates from each positive faecal sample for serogrouping. In previous research conducted in our laboratory, Salmonella isolates cultured from the same faecal sample and belonging to the same serogroup, were often identified as the same serotype [Reference Edrington7, Reference Fitzgerald12]. Therefore in the current research, each isolate identified from an individual faecal sample and belonging to the same serogroup were assumed to be of the same serotype. Each isolate identified as belonging to a different serogroup were serotyped and the results presented in Table 6. This data highlights the serotype diversity and shifting prevalence within individual animals over time and demonstrates the difficulty in determining the optimum number of isolates required for serotype prevalence determination. One animal yielded four and another animal five different serotypes from one Salmonella-positive faecal sample (Table 7).
Table 7. Salmonella serotype distribution by month for individual healthy dairy cows (2004). Five isolates per positive faecal sample serogrouped and only different serogroups sent for serotyping. Isolates identified as belonging to the same serogroup were assumed to be the same serotype

Neg., Salmonella-negative faecal sample; n.s., no sample.
DISCUSSION
In an attempt to elucidate potential contributing factors to salmonellosis outbreaks in mature lactating dairy cattle, we examined isolates collected from diarrhoeic and healthy cattle during an outbreak in 2003. While the number of different serogroups and serotypes was greater in sick compared to healthy cattle, no differences between the two groups were particularly noteworthy with one exception. The serotypes Kentucky and Senftenberg were identified only in the sick cattle, with their respective serogroups accounting for a significant portion of the overall prevalence (19% and 21%, respectively) in comparison to the other serogroups identified. However, we have previously reported these serotypes in apparently healthy lactating dairy cattle [Reference Edrington7, Reference Fitzgerald12] and cannot attribute the salmonellosis observed in 2003 solely to these serotypes. Others have also reported isolating Salmonella Kentucky from healthy [Reference Wells3, Reference Huston16–18] and diarrhoeic [Reference Sato9] dairy cattle. Serotype Barranquilla was also identified only in the sick group, however, only one cow had that particular serotype and serogroup. It seems likely that if Barranquilla isolates were the causative agent of illness, we would have identified that serogroup in other sick cattle.
Further analysis of the Salmonella isolates from sick and healthy cattle was conducted using PFGE. While multiple genotypes were identified within serotype, no distinguishable differences were observed when comparing sick vs. healthy isolates. Six different genotypes were observed within the Montevideo serotype out of 22 isolates examined. The Mbandaka and Cubana serotypes each contained three different genotypes while all nine isolates of the serotype Soerenga were the same genotype. These results are similar to previous research reported by the authors in which multiple genotypes were observed for serotypes in which multiple isolates were examined including Montevideo, Mbandaka, and Cubana, however, as in this research the one exception was the serotype Soerenga, where only two genotypes were identified from 11 isolates [Reference Edrington7].
We did not examine the faecal samples in 2003 for any pathogens other than Salmonella and relied on the attending veterinarian's diagnosis of salmonellosis being correct. Coupled with our examination that failed to identify any distinguishing differences among Salmonella isolates from sick and healthy cattle, it is possible that other pathogens may be responsible or contributing to the outbreak. However, similar outbreaks have occurred in these dairies previously and were diagnosed as salmonellosis. Unfortunately, at the time we did not quantify Salmonella concentrations in the faecal samples and in light of the current results, feel that this may have yielded more useful information that supports our hypothesis for the outbreak discussed below.
The following year we collected monthly samples from the same dairy that experienced the 2003 salmonellosis outbreak. Thirty cows all calving within 2 weeks of each other were sampled on a monthly basis over a 9-month period that incorporated the time-frame when salmonellosis outbreaks had previously occurred (August and early September). Water (from the trough), soil and TMR samples were collected, representative of the pens housing the cattle over the course of the collection period. Fortunately for the producer, the cows did not experience an outbreak of salmonellosis during 2004. The increase in overall faecal prevalence in June compared to May, and in August compared to July (particularly faecal prevalence), led us to believe that perhaps conditions were changing to favour an outbreak, but in September and October we observed a sharp decline in faecal prevalence as well as a shift in faecal serogroups.
Salmonella can survive for prolonged periods in the environment potentially contributing to chronic herd infection [Reference Gay and Hunsaker8, Reference Giles19, Reference Wray20], therefore it was not surprising to isolate this organism from the pen soil and water samples. Somewhat unexpected, however, is the lack of a common serogroup between faecal samples and potential routes or infection or re-infection, namely feed, water and soil. In some months the same predominant serogroup was observed for all sample types, while in other months, serogroup varied with sample type. The diversity of serogroups observed in TMR samples is unexpected. Salmonella has been reported in animal and plant protein sources and by-product feeds commonly fed to dairy cattle [Reference Franco21–Reference Wray and Sojka23] with a report that 40% of the protein animal by-product sources in the United States were contaminated with Salmonella [Reference Smith and Smith24]. However, while we attempted to collect samples that had not been disturbed by the cattle, we cannot rule out this as possible source of contamination. Similarly, insects, birds, and rodents are all possible contamination sources. Additionally, water was added to the TMR prior to feeding and may have served as a Salmonella source. In the second half of the experimental period, we collected water directly from the taps providing water to the troughs as well as the trough water. Only one tap sample was Salmonella positive which suggests that water may not have been the predisposing TMR contaminant source. Beginning on 23 March all drinking water was chlorinated on the farm as a means of Salmonella control. Due to the hardness of the water in this region, consistent chlorination was not achieved and the effectiveness of this control measure was difficult to determine. Possibly the chlorination was partially effective as only one tap-water sample was Salmonella positive, whereas continuous water trough contamination from cattle and other sources may have been sufficient to overwhelm the effectiveness of chlorine.
Results of the monthly collections highlight the extreme variation in Salmonella prevalence as well as serogroup and serotype diversity within a population of dairy cattle. We were not able to find any correlation of serogroup or serotypes in healthy or diarrhoeic cattle. Furthermore, we identified multiple serotypes from the same faecal sample on a number of occasions and have reported similar results previously [Reference Fitzgerald12]. In one animal, all five randomly selected isolates from a single Salmonella-positive faecal sample were identified as five distinct serotypes. Others have reported exactly the opposite. Wells and colleagues reported 12% of the dairies sampled had multiple serotypes, although culture-positive cows frequently had only one Salmonella serotype isolated [Reference Wells17]. In the San Joaquin Valley of California, Salmonella was cultured in 16% of the dairies, however, not more than one serotype was found on any one dairy [Reference Pacer25]. Contrary to these reports and consistent with our research, the isolation of multiple Salmonella serovars was reported in a single California dairy [Reference Gay and Hunsaker8]. Serotype diversity was reported to be greater for samples collected in the western United States compared to other locations [Reference Galland26, Reference Troutt27].
Asymptomatic Salmonella carriage and faecal shedding in adult dairy cattle is fairly common [Reference Edrington6, Reference Edrington7, Reference Huston16, Reference Wells17], with salmonellosis typically associated with young calves. Clinical cases of salmonellosis have been reported in adult dairy cattle and the causative agent speculated [Reference Gay and Hunsaker8–Reference Anderson10]. Feed ingredients, diet changes, other infections and stress have all been suggested as potential contributors to salmonellosis in adult dairy cattle [Reference Anderson10, Reference Wray and Sojka23, Reference Moore28, Reference Van Dreumel29]. We have sampled this dairy and others in the region on multiple occasions and frequently isolated Salmonella from faecal samples. As multiple serotypes have been isolated in these animals and based on the comparison of healthy vs. sick isolates, it is unlikely that a single serotype is responsible for the outbreak of salmonellosis. Over 2400 different Salmonella serotypes have been identified and all can be considered pathogenic depending on exposure level and host resistance [Reference Wells3]. A more plausible explanation may be achieved by examining the average monthly temperature data for the region. Ambient temperatures increase throughout the spring and summer, peaking at approximately the same time that previous salmonellosis outbreaks have been reported. Although measures to alleviate heat stress (e.g. shade, water misters) are employed by the farm, heat stress is an inherent part of dairy production in this region of the United States. We speculate that the salmonellosis outbreaks are a result of chronic heat stress that sufficiently weakens the animal's defence system to allow a rapid increase in overall Salmonella populations (not serotype specific) within the gastrointestinal tract thereby causing infection. Future outbreaks will include quantification of Salmonella populations throughout the gastrointestinal tract as well as culture for other pathogens that may be responsible in whole or part for the observed symptoms.
The research reported herein highlights the complexity of Salmonella control at the farm level. While reducing Salmonella prevalence on the farm has important cow health, environmental and food safety implications, this remains a daunting task. Previously we have attempted to identify potential factors contributing to Salmonella shedding that might pinpoint times or groups of cattle where implementation of Salmonella control measures would be most effective. Our research to date indicates that any on-farm control programmes will need to have broad reaching effects, encompassing animals at all ages and stages of production as well as the dairy environment.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the excellent technical expertise of Kate Andrews and Reiley Street.
DECLARATION OF INTEREST
None.












