FFQ are often used to determine diet–disease relationships in epidemiological research because they are inexpensive and pose a low burden on participants compared with other dietary assessment methods. However, the association between disease and dietary exposure, assessed by an FFQ, is biased because of measurement errors in the FFQ( Reference Kipnis, Subar and Midthune 1 ). Therefore, a validation study should be performed to assess the amount of measurement error in order to correct the observed associations. However, whether the correction is adequate depends among others on the characteristics of the reference method used in the validation study. A reference method should (1) be unbiased and (2) have uncorrelated errors with the errors in the method to be validated( Reference Kaaks, Riboli and Esteve 2 ). Recovery biomarkers are assumed to meet these requirements, but are only available for energy and for a few nutrients such as K, Na and protein( Reference Jenab, Slimani and Bictash 3 , Reference Willett 4 ). Therefore, other dietary assessment methods such as replicate 24 h recalls (24hR) and food records have been used as reference methods. However, previous research showed that these methods do not entirely correct for measurement errors( Reference Kipnis, Subar and Midthune 1 , Reference Ferrari, Roddam and Fahey 5 – Reference Geelen, Souverein and Busstra 7 ) because they are biased and have correlated errors with the FFQ.
Bias is present when dietary intake is over- or under-estimated because of, for example, incorrect portion-size estimation, inaccuracies in food composition databases (FCD) or a lack of detail to identify foods consumed. The second criterion for a valid reference method, that is, uncorrelated errors between the reference method and FFQ, is violated when, for example, both methods make use of data from the same FCD, rely on memory or estimate portion sizes by using the same household measures( Reference Willett 4 ). The duplicate portion (DP) technique partially overcomes these limitations as it does not depend on FCD data, is not memory based and does not use standardised portion sizes. For a DP, participants collect a second equal portion of each food and drink they consume over 1 or more days. Afterwards, the dietary composition of the DP is determined by chemical analysis. Because of this, the magnitude of correlated errors of this method with an FFQ is expected to be lower than that of a 24hR for which correlated errors are a known limitation( Reference Kipnis, Subar and Midthune 1 ). On the other hand, collections of DP may lead to reactivity bias, demonstrating a change in the respondents’ intake on the collection day, mostly resulting in underestimation of intake( Reference Isaksson 8 – Reference Stockley 11 ).
Our study aimed to evaluate the suitability of the DP technique as a reference method for an FFQ to assess protein, K and Na, using multivariate measurement error models. As the 24hR is often used as a reference method in evaluation studies, our secondary aim was to compare the validity of the 24hR and DP as reference methods for an FFQ. To this end, recovery biomarkers for protein, K and Na were determined and assumed to be unbiased with independent measurement error.
Methods
Subjects and design
In this study, the DuPLO study, a random subsample of 200 Dutch adults (ninety-two men and 108 women) from the NQplus study were included. The NQplus study is a longitudinal study designed for multiple aims: to validate a newly developed FFQ; to start a reference database for nutrition research; and to study the association between diet and intermediate health outcomes. Participants for the NQplus study were recruited by sending invitations to randomly selected persons aged 20–70 years, living in Wageningen, Ede, Renkum and Arnhem. Subjects participating in the NQplus study at that time (n 630) received an email invitation to join the DuPLO study. Recruitment for DuPLO started in November 2011 until April 2013. After reaching the intended sample size for DuPLO (n 200), recruitment for the NQplus study was still ongoing.
Baseline measurements consisted of, among others, a physical examination, including weight and height, and general and lifestyle questionnaires (including questions about education, health and smoking habits). Within a timeframe of 3 years each participant collected two DP (approximately 5 months apart) and two urine samples (approximately 1 year apart). In addition, two self-reports by FFQ (approximately 7 months apart) were handed in. The 24hR was administrated in two ways: by means of a telephone interview by a trained dietitian (telephone-based 24 h recall collection (24hRT)) (0–8 replicates, approximately 4 months apart) or filled in by the participant in a web-based programme (web-based 24 h recall collection (24hRW)) (0–9 replicates, approximately 3 months apart). An overview of the timeframe and sample size of the data collection is presented in Appendix I. The large variety in replicates for the 24hRT and 24hRW is mainly due to the fact that participants were difficult to reach by telephone or people felt that the burden of participation was too much and therefore cancelled invitations for the 24hR. The Dutch FCD of 2011( 12 ) was used to calculate nutrient intake for the 24hRT, 24hRW and FFQ. Participants with missing data for one or more of the methods were included in the analysis because they provided information for the other dietary assessment methods. In total, 198 participants were included for analysis: ninety-two males and 106 females. Two participants became pregnant during the study. As it was expected that they deviated from their habitual dietary intake, they were excluded from analysis. This study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all procedures involving human subjects were approved by the medical ethical committee of Wageningen University. Written informed consent was obtained from all subjects.
Dietary assessment
Duplicate portion collection
Participants received verbal and written instructions to collect a second identical edible portion of all foods and drinks consumed over a 24 h period. Foods and drinks were collected in separate baskets in a cool box (5°C). Participants received a monetary reimbursement for the products collected for the DP. The collected and consumed portions were measured using the same household measures. The collection cool boxes were brought to the participant’s home 1 d before collection and picked up the day after collection. In the laboratory, collected DP were weighed, homogenised in a blender (Waring Commercial model 34BL22; Waring) and 2·5 ml 0·02 % tert-butylhydrochinon (BHQ) in ethanol was added per kg of DP as antioxidant during blending. The homogenised samples were stored within 1 h at –20°C until further analysis. A part of the sample was freeze-dried before analysis.
FFQ
Participants completed an online self-administered 180-item FFQ using the online open-source survey tool LimesurveyTM (LimeSurvey Project Team/Carsten Schmitz, 2012). Portion sizes were assessed by commonly used household measures, and the reference period for reporting intake was the past month. The performance of the FFQ had been evaluated for energy (ρ=0·65 as compared with three 24hR), fats (ρ ranged between 0·29 and 0·75 as compared with three 24hR), selected vitamins (ρ ranged between 0·46 and 0·86 as compared with three 24hR) and dietary fibre intake (ρ=0·82 as compared with three 24hR)( Reference Streppel, De Vries and Meijboom 13 ). The estimated mean energy intake by the FFQ appeared to be accurate( Reference Siebelink, Geelen and De Vries 14 ), and in comparison with a replicate 24hR the FFQ showed an acceptable to good ranking ability for most nutrients( Reference Streppel, De Vries and Meijboom 13 ).
Web-based 24 h recall collection
Participants received an unannounced email invitation, which was valid for 24 h, to self-administer a recall over the previous day in the web-based programme Compl-eat. This programme is based on the five-step multiple-pass method( Reference Conway, Ingwersen and Vinyard 15 ), which enables participants on a step-by-step basis to accurately report the foods and drinks consumed the previous day. If participants did not fill in the 24hRW, a new invitation was randomly sent within 3–10 d. Portion sizes of foods or recipes were reported by using household measures, standard portion sizes, weight in grams, or volume in litres( Reference Donders-Engelen and van der Heijden 16 ). The 24hRW were checked for completeness and unusual or missing values, and, if necessary, adjustments were made using standard portion sizes( Reference Donders-Engelen and van der Heijden 16 ) and recipes following a standard internal protocol.
Telephone-based 24 h recall collection
Trained dietitians of the Division of Human Nutrition of Wageningen University made an unannounced phone call to the participant. The dietitian asked about foods and drinks consumed the previous day according to a standardised protocol based on the five-step multiple-pass method( Reference Conway, Ingwersen and Vinyard 15 ). The 24hRT were coded using Compl-eat. For various components (energy, nutrients and foods) the highest and lowest ten values were checked for errors, such as errors in coding numbers or in the amounts (e.g. 150 cups instead of 150 g of milk).
Urine collection
Participants received verbal and written instructions for 24 h urine collections. The urine collection started after discarding the first voiding on the morning of the collection day and finished after the first voiding on the morning of the next day. The preservative lithium dihydrogenphosphate (25 g) was added to the collection containers. Subjects were instructed to ingest a tablet containing 80 mg para-aminobenzoic acid (PABA) (PABA check; Elsie Widdowson Laboratory) during breakfast, lunch and dinner on the day of collection to check for completeness of urines. Participants were also instructed to register possible deviations from the protocol (e.g. missing urine). At the study centre, the urine collections were mixed, weighted and aliquoted and stored at –20°C until further analyses.
Laboratory analysis
Protein analysis
Total N in the urine and in the DP was analysed by the automated Kjeldahl method( Reference Hambleton and Noel 17 ) using a Foss KjeltecTM 2300 Analyzer (Foss Tecator AB). The amount of protein was calculated using a N-to-protein conversion factor of 6·25( Reference Jones 18 ). Protein intake was calculated from N excretion, assuming an average ratio of urinary-to-dietary N of 0·81( Reference Bingham and Cummings 19 ). For the DP, the within-run CV was <1 % and between-run CV was <1 %. For the urine analysis, the within-run CV was 1·6 % and between-run CV was 1·3 %.
Potassium and sodium analysis
K and Na in urine were determined with an ion-selective electrode (Roche 917 analyser; Roche). K and Na intake assessed by urinary excretion was calculated taking into account 19 % K( Reference Freisling, Van Bakel and Biessy 20 ) and 14 % Na( Reference Holbrook, Patterson and Bodner 21 ) extra-renal and faecal losses. Participation in the External Quality Assessment Scheme of the Dutch Foundation for Quality Assessment in Medical Laboratories showed a bias of –1·6 % and +1·1 %, and the analytical variation was 1·6 % and 1·2 % for K and Na, respectively. A within-run CV of <1 % and a between-run CV of <1 % for K and a within-run CV of <1 % and a between-run CV of <1 % for Na were observed. K and Na in the DP were determined after digestion of the samples in PTFE tubes using a MarsXpress microwave digestor (CEM), with inductively coupled plasma atomic emission spectroscopy (ICP-AES, Varian Australia Pty Ltd, ISO, 2010) at the Chemical Biological Soil Laboratory of Wageningen University with a within-run CV of <1 % and a between-run CV of <1 % for K and a within-run CV of 1·1 % and a between-run CV of 1·7 % for Na.
Para-aminobenzoic acid analysis
PABA was measured by means of HPLC after alkaline hydrolysis of the urine samples to convert PABA metabolites into PABA( Reference Jakobsen, Ovesen and Fagt 22 ). Using a minimum of 78 % PABA recovery as a cut-off point for complete urine collection, which is proposed if PABA is analysed by HPLC( Reference Jakobsen, Ovesen and Fagt 22 ), 16·7 % of the urine samples were judged incomplete. The total CV for the PABA analysis was 9 %. The within-run CV for PABA was 1·9 %, and the between-run CV for PABA was 1·3 %.
Measurement error model
We assumed protein, Na and K intake assessed by urinary excretion to be unbiased in assessing usual intake( Reference Jenab, Slimani and Bictash 3 ), which we assumed not to vary within the 3 years of study. All our measurement error models assumed a linear relationship between DP, 24hRT, 24hRW, FFQ, biomarker and the true unknown intake T. In our measurement error model i is the person, and j indicates the occasion. Furthermore, α X expresses the constant bias for reference method X (X being DP for the DP method, 24hRT for the telephone-based 24hR, and 24hRW for the web-based 24hR) and β X is the proportional scaling bias where α Q and β Q are similar respective parameters for the FFQ. The person-specific bias of the reference method is given by w xi and for the FFQ by v i . Finally, ε Xij is the random error with mean zero and constant variance for the reference method, whereas ε Qij is the random error for the FFQ.
Statistical analysis
Descriptive statistics were presented in percentages and as means with their standard deviation. Presence of bias between the mean of the recovery biomarker and the mean of the available replicates of FFQ, DP, 24hRW and 24hRT was tested by performing a Student’s paired t test. The significance level was set at a two-sided P value of 0·05.
A Bayesian approach( Reference Richardson and Gilks 23 ), Markov Chain Monte Carlo, the PROC MCMC procedure in SAS, was used to estimate the parameters of our measurement error models for which uninformative priors were set to make the model data driven (syntax can be found in Appendix II). The sensitivity of our measurement error model was tested by using different distributions for the parameters and changing the prior estimates. As little variation in model outcomes was observed, we assumed the model to be robust. Sex-specific models for Na did not converge (because of the low variance of the person-specific biases compared with within- and between-person variances) and are therefore not reported. To assess whether the reference method adequately corrects for measurement error it should be unbiased, which is indicated by the absence of proportional scaling bias (a β x equal to one in equation 1 of the measurement error model indicates that there is no proportional scaling bias present). Furthermore, the reference method should have uncorrelated errors with the errors in the FFQ; that is, the error correlation should be 0. The error correlation (ρ XQ ) is calculated according to formula 4 specified below from the measurement error model outcomes. From the model outcomes we also calculated the attenuation factor (λ X ) for each reference method according to formula 5 as specified below. Note that this is not the attenuation factor for the FFQ using the reference method, but the attenuation factor for the reference method using the biomarker as reference.
where cov wivi is the covariance between the error in the FFQ and the error in the reference method X; varε Xij is the variance of the random error of the reference method X; varw xi indicates the variance of the person-specific bias of method X; varv i is the variance of the person-specific bias of the FFQ; varε Qij is the variance of the random error of the FFQ; and β X is the proportional scaling bias of method X. To obtain the estimates of the attenuation factor for multiple DP and 24hR, the variance of the random error of the method (varε Xij ) was divided by the number of measurements (k) of the method. All statistical tests were performed in SAS version 9.3 (SAS Institute Inc., 2012).
A sensitivity analysis was performed, comparing the model outcomes from the complete urine data set with the model outcomes after exclusion of the urine samples with <78 % PABA recovery( Reference Jakobsen, Ovesen and Fagt 22 ). Measurement error model outcomes did not differ substantially when no urine samples were excluded compared with excluding urines with PABA <78 %. This points in the same direction as the finding of Subar et al. ( Reference Subar, Midthune and Tasevska 24 ), who observed a modest effect on correction factors when urines were excluded on the basis of PABA recovery compared with not excluding urines in the OPEN study( Reference Subar, Midthune and Tasevska 24 ). We therefore report the results based on the complete urine set in this article.
Results
At baseline, participants were on average 55·7 (sd 10·2) years of age, and women were slightly younger than men (53·8 v. 58·0 years, Table 1). The average BMI was 25·1 (sd 3·7) kg/m2, and a higher percentage of women (64 %) had a healthy BMI (18·5–25·0 kg/m2) compared with men (46 %). Furthermore, 58 % of the men and 48 % of the women were classified as highly educated (university or college).
Table 1 Baseline characteristics of the study population (Mean values and standard deviations; percentages)
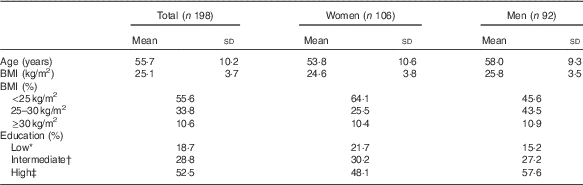
* Primary or lower education.
† Secondary or higher vocational education.
‡ University or college.
The percentage of the number of 24hRT and 24hRW varied between 18 and 29 % over the seasons (Table 2). The variation in the number of urine collections per season was larger, and varied between 4 % collected in spring and 51 % in summer. Most DP (34 %) were collected in spring (Table 2). For the FFQ, 39 % were collected in autumn and 12 % in winter. The DP, 24hRT, 24hRW and urine collections were evenly distributed between week (range, Monday–Friday 63–76 %) and weekend days (range, Saturday–Sunday 24–37 %).
Table 2 Percentage of the number of collection days distributed over the seasons and weekend v. weekdays
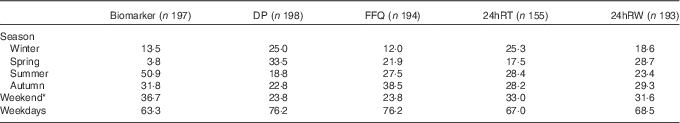
DP, duplicate portion; 24hRT, telephone-based 24 h recall; 24hRW, web-based 24 h recall.
* Weekend days are Saturdays and Sundays.
The DP underestimated protein by 20·9 %, K by 6·8 % and Na by 33·5 % (Table 3). For all nutrients, underestimation was smallest using the 24hRT (protein 12·7 %, K 4·7 % and Na 28·7 %). The FFQ, the method to be validated, underestimated protein (22·6 %) and Na (41·6 %) to the largest extent. A similar pattern was observed for men and women. Overall, women tended to underestimate to a lesser extent than did men for all dietary assessment methods and nutrients.
Table 3 Mean intake and bias for the intake of protein, potassium and sodium, compared with the urinary excretion marker (Mean values and standard deviations)

n, number of participants.
* Values were significantly different from the biomarker (P<0·01).
† For Na, sex-specific models did not converge because of the low variance of the person-specific biases compared with within- and between-person variances and are therefore not reported.
A proportional scaling bias, as indicated with β x in Table 4, closer to 1 means less bias. In general, the estimates for the DP were closest to 1, 0·58 for protein, 0·72 for K and 0·52 for Na, compared with those for 24hRT and 24hRW (Table 4). For the sex-specific models, the proportional scaling bias was closest to 1 for the DP for K for women (0·77) and for protein for men (0·72). However, the 24hRT performed better for protein for women (0·62) and for K for men (0·93).
Table 4 Proportional scaling bias and correlated error with the FFQ for the intake of protein, potassium and sodium (Mean values and standard deviations)

24hR, 24 h recall.
* Adjusted for BMI and sex.
† Adjusted for BMI.
‡ For Na, sex-specific models did not converge because of the low variance of the person-specific biases compared with within- and between-person variances and are therefore not reported.
In the total population, the correlated errors between the DP and FFQ were the lowest for the two micronutrients, Na (0·19) and K (0·17) (Table 4). For protein, the error correlations with the FFQ were comparable between the three reference methods (0·28 for the DP and 24hRT, and 0·27 for the 24hRW). The range of correlated errors was comparable for men (0·12–0·28) and women (0·08–0·29).
An attenuation factor close to one indicates an overall better estimation of the nutrient intake. In the total population, looking at estimates for single measurements, attenuation factors for the DP were highest for all three nutrients (0·74 for protein, 0·54 for K and 0·43 for Na), whereas for the 24hRW attenuation factors tended to be the lowest for all nutrients (0·30 for protein, 0·31 for K and 0·18 for Na) (Table 5). The same trend was seen for women and men separately. Attenuation factors increased when the number of replicates was expanded. For protein, the attenuation factor for one measurement of the DP was 0·74, whereas for the 24hRT three measurements gave a similar attenuation factor (0·73). In general, attenuation factors for all dietary assessment methods tended to be higher for men than for women.
Table 5 Attenuation factors for the reference methods for the intake of protein, potassium and sodium (Mean values and standard deviations)

DP, duplicate portion; 24hRT, telephone-based 24 h recall collection; 24hRW, web-based 24 h recall collection.
* Adjusted for BMI and sex.
† Adjusted for BMI.
‡ For Na, sex-specific models did not converge because of the low variance of the person-specific biases compared with within- and between-person variances and are therefore not reported.
Discussion
In this Dutch validation study, we found that all dietary assessment methods underestimated the intake of protein, K and Na compared with the biomarker measurements where the 24hRT showed the smallest underestimation. Furthermore, all dietary assessment methods were biased (affected by proportional scaling bias) and showed correlated errors with the FFQ for protein, K and Na. However, dietary intake measures from the DP were less affected by proportional scaling bias compared with the 24hRT and 24hRW. Furthermore, error correlations between the DP and FFQ were the lowest. Attenuation factors also indicated that the DP had the best performance (attenuation factors were closer to one).
To our knowledge, this is the first study assessing error correlations between the FFQ and DP, proportional scaling bias for DP and estimating attenuation factors for the DP. Research on 24hR has among others been performed in a pooled analysis of five American validation studies comparing protein intakes assessed by the FFQ and 24hR with urinary N excretion( Reference Freedman, Commins and Moler 25 ). Freedman et al. ( Reference Freedman, Commins and Moler 25 ) found wide ranges of study-specific attenuation factors (0·14–0·54) for the 24hR. This is comparable to our results, but we found estimates at the higher end of this range. One of the possible explanations is that our study population was highly motivated; they were willing to collect, in addition to filling out multiple 24hR and various food and lifestyle questionnaires, two urine and two DP samples. Above that, a high percentage of our participants were highly educated. Furthermore, cultural differences in dietary patterns and the design of the FFQ and 24hR could also explain our findings to be in the upper part of the range.
Proportional scaling bias for the 24hRT for protein was similar to that found in the OPEN study, a large American study from Montgomery County, Maryland, for women (0·62 for DuPLO v. 0·60 for OPEN), but our estimate was slightly lower for men (0·64 for DuPLO v. 0·70 for OPEN)( Reference Kipnis, Subar and Midthune 1 ). Error correlations between the 24hR and FFQ were slightly higher in our study compared with the EPIC study, a large European multi-centre study, showing 0·21 for K and 0·21 for protein( Reference Ferrari, Roddam and Fahey 5 ), and the OPEN study (showing 0·24 for protein for women and 0·18 for men)( Reference Kipnis, Subar and Midthune 1 ). Prentice & Huang( Reference Prentice and Huang 26 ) found slightly higher error correlations between their FFQ and 24hR for protein (0·33)( Reference Prentice and Huang 26 ). Differences between error correlations of the 24hR with the FFQ in studies are expected because of different sets of covariates included – different modes of administration (web based and interviewer administered) and numbers of replicates of a 24hR – varying ways of portion-size estimations and differences between the study populations (ethnic groups, social economic status, age).
The attenuation factor for Na intake for the DP (0·43) was remarkably higher than for both 24hR administrations (0·19 for the 24hRT and 0·18 for the 24hRW), and taking a second replicate for the DP increased the attenuation factor to 0·65. The DP for Na was also less affected by proportional scaling bias (β DP=0·52) and demonstrated a lower error correlation with the FFQ (0·19) compared with the 24hRT and 24hRW. Accurately assessing Na intake is challenging because of the high variability of Na content of foods( Reference Champagne and Cash 27 ), which is not always accurately reflected in FCD. In addition, it is difficult to accurately report the amount of salt added during cooking or at the table. In the 24hR and FFQ in this study, there is no question included about added salt during cooking or at the table. The accuracy of dietary intake estimates of Na from 24hR and FFQ is therefore expected to be limited. This is supported by other research about Na estimation from 24hR, FFQ and dietary records( Reference Champagne and Cash 27 ). The higher attenuation factor and proportional scaling bias for the DP could be explained by the fact that salt added during cooking was included as a sample of the cooked meal was collected and the DP were chemically analysed and estimates did not depend on information in FCD. However, attenuation factors for Na for the DP were still notably lower than those for protein and K intake.
Correlated errors between the FFQ and reference methods for protein intake tended to have the same order of magnitude for all methods, whereas for K and Na intake the DP showed lower error correlations than the 24hRT and 24hRW. Thus, there must be a source of error equally influencing the estimation of protein in all four methods apart from the correlated errors that are expected between the FFQ and 24hR (use of the same FCD to calculate nutrients, estimation of portion sizes and memory based). A similar error source for all four methods (FFQ, DP, 24hRT and 24hRW) could be response errors, meaning that people tended to forget (for FFQ and 24hR) or not collect (for DP), either on purpose or not, protein-rich products.
A weakness of this study is the unequal spread of biomarker measurements over the seasons (summer was over-represented and spring under-represented), while they were assumed unbiased in our measurement error model. This assumption was based on evidence from the literature that does not indicate seasonal variation of nutrient intake in western populations( Reference Ma, Olendzki and Li 28 , Reference van Staveren, Deurenberg and Bureman 29 ). Furthermore, the different methods did not exactly cover the same time period. However, we were interested in a person’s usual intake and not in the dietary intake on a specific day. We assumed that energy and nutrient intake of a person would be fairly stable over a longer time period. Thus, although intake data measured by the different dietary assessment methods did not cover the same time period, they could be all considered to represent a person’s usual energy and nutrient intake. Therefore, comparisons between methods can be made.
We reported the results based on all urine samples collected, independent of the PABA results. This was based on a sensitivity analysis to exclude urine samples based on PABA, focussing on the main outcomes: attenuation factor and correlated error. These main outcomes did not differ substantially between inclusion of all urine samples and inclusion of only the complete urine samples (based on PABA recovery). Furthermore, not excluding urine samples provided a larger sample size. However, results for bias (i.e. difference between levels of intake) must be regarded rather carefully as they differed significantly for protein and K when incomplete urine samples were excluded.
Taking into account that in general the DP showed lesser proportional scaling bias, the highest attenuation factors and the lowest error correlations with the FFQ, this method appeared more promising as a reference method than did the 24hR. Important considerations in the collection of DP are that it is burdensome for participants, requires a lot of time from the researcher, is expensive to perform and reactivity bias – mostly causing underestimation of habitual intake – is expected. We carefully instructed our participants not to deviate from their habitual intake and provided them with written instructions, including tips to remind the participant to include everything in the collection baskets. Nevertheless, the DP showed substantial underestimation for protein, K and Na.
Attenuation factors calculated for FFQ using the 24hR as a reference method are affected by correlated errors between the two methods( Reference Wong, Day and Bashir 30 ). Better estimates of attenuation factors will be obtained if these correlated errors between the FFQ and 24hR are taken into account. The error correlations between the 24hR and FFQ found in this study could be considered in the calculation of attenuation factors; however, generalising results from one study population to another should always be done conservatively, taking into account the characteristics of both study populations and the study setup.
Conclusion
We conclude that the DP violated the requirements to be used as unbiased reference method for validating an FFQ, however, to a lesser extent than a telephone-based 24hR and, even more, a web-based 24hR. As the proportional scaling bias was less for the DP, the DP-FFQ error correlations were lowest, and the attenuation factors were highest, we propose that the DP is probably the best available reference method for FFQ validation for nutrients that currently have no generally accepted recovery biomarker.
Acknowledgements
The authors thank Professor Edith Feskens and Anne van de Wiel, MSc, for making it possible to use data from the NQplus study. The authors also thank Mira Mutiyani, BSc, Sanne Marije Seves, BSc, and Cecilia Ferreira Lima, BSc, for their help in analysing the duplicate portion samples and Corine Perenboom for her help in preparing the 24 h recall data. In addition, the authors thank the subjects of the DuPLO study for participating in this study.
The NQplus study was funded by ZonMw (grant no. 91110030) and Wageningen University. The DuPLO study was funded by VLAG (Voeding, Levensmiddelentechnologie, Agrobiotechnologie en Gezondheid), a graduate school of Wageningen University. The sponsors had no role in study design, analysis and interpretation of the data or in writing of the article.
The authors’ contributions are as follows: L. T. collected the data and contributed to the study design, data analysis and interpretation of findings and wrote the manuscript. J. H. M. d. V, P. v. V. and A. G. contributed to the study design, interpretation of findings and revised the earlier versions of the manuscript. H. C. B. contributed to the data analysis, interpretation of findings and revised the earlier versions of the manuscript. P. J. M. H. and P. C. H. H. contributed to the study design and revised the earlier versions of the manuscript. All authors read and approved the final version of the manuscript.
There are no conflicts of interest.
Appendix I: overview of timeframe and sample size of the data collection

Appendix II: Syntax for the MCMC procedure
In this appendix, we provide the SAS syntax for our measurement error model using proc MCMC. We used generic labels for the variables as can be found in the model statement. The data set is called mydata. The array statement is used for identifying the (latent) person-specific biases and the error covariance. In the parms statements, the starting values are given for each model parameter, and in the prior statement a distribution is given. The estimates for the correlated errors and attenuation factors are calculated at the bottom of the syntax.
ods graphics on;
Proc MCMC data=mydata seed=20000 nmc=300000 thin=20 NBI=50000 Maxtune=50 MONITOR=(_PARMS_ corrDPQ rhoDPT lambdaDP rho2DPT lambda2DP rhoQT lambdaQ) outpost=postdata;
ARRAY WIVI[2] VI wi;
array wivi_0[2] (0,0);
ARRAY varwivi[2,2];
array s[2,2] (1 0 0 1);
parms bDP 0·5 aDP 76 sdEDP 10;
parms aQ 71 bQ 0.5 sdEQ 10;
parms muT 100 sdT 25 sdEM 15;
parms bBMI1 0 bBMI2 0;
parms bG1 0 bG2 0;
parms varwivi {1 0 0 1};
parms wiscale 15;
parms viscale 25;
prior aQ~normal (0, var=10000);
prior bQ~normal (0, var=10000);
prior aDP~normal (0, var=10000);
prior bDP~normal (0, var=10000);
prior bBMI1~normal (0, var=10000);
prior bBMI2~normal (0, var=10000);
prior bG1~normal (0, var=10000);
prior bG2~normal (0, var=10000);
prior muT~normal (0, var=1000000);
prior sdEQ~uniform (0, 50);
prior sdEDP~uniform (0, 50);
prior sdEM~uniform (0, 50);
prior sdT~uniform (0, 50);
prior viscale~uniform (0, 50);
prior wiscale~uniform (0, 50);
prior varwivi~iwish (3,s);
varEQ=sdEQ**2; varEDP=sdEDP**2; varEM=sdEM**2; varT=sdT**2;
Random T~normal (muT, var=varT) subject=_OBS_;
Random wivi~mvn (wivi_0, varwivi) subject=_OBS_;
muQ=aQ+bQ*(T-100)+bG1*gender+bBMI1*(BMI-25)+viscale*vi;
muDP=aDP+bDP*(T-100)+bG2*gender+bBMI2*(BMI-25)+wiscale*wi;
muM=T;
model FFQ_1_prot~normal (muQ, var=varEQ);
model FFQ_2_prot~normal (muQ, var=varEQ);
model DP1_prot_gr~normal (muDP, var=varEDP);
model DP2_prot_gr~normal (muDP, var=varEDP);
model T0_urine_eiwit~normal (muM, var=varEM);
model T1_urine_eiwit~normal (muM, var=varEM);
corrDPQ=wiscale*viscale*varwivi[1,2]/sqrt(((wiscale**2)*varwivi[2,2]+varEDP)*((viscale**2)*varwivi[1,1]+varEQ));
lambdaDP=(1/(1+(varEDP+(wiscale**2)*varwivi[2,2])/(bDP**2*varT)))/bDP;
lambda2DP=(1/(1+((varEDP/2)+(wiscale**2)*varwivi[2,2])/(bDP**2*varT)))/bDP;
run;
ods graphics off;








