Introduction
The Agromyzidae (Diptera) is a family of small flies which has a significant economic impact on agricultural and horticultural crops around the world. The larvae of many species mine leaves, but some species mine stems or form galls (Spencer, Reference Spencer1973). The most well-known genus of this family is Liriomyza, which contains over 400 species; however, only a few species are considered destructive pests globally (Parrella, Reference Parrella1987). Most plant damage is caused by the larvae tunneling within the mesophyll of leaves, leaving serpentine or blotch mines (Parrella et al., Reference Parrella, Jones, Youngman and Lebeck1985). Female flies can also damage plants by using their ovipositors to penetrate the epidermis of leaves, creating numerous punctures for feeding and ovipositing (Bethke and Parrella, Reference Bethke and Parrella1985; Ge et al., Reference Ge, Wei, Tao and Kang2019). Infested plants usually have reduced photosynthetic rates and young seedlings can die when very heavily mined (Johnson et al., Reference Johnson, Welter, Toscano, Ting and Trumble1983; Bueno et al., Reference Bueno, Zechmann, Hoback, Bueno and Fernandes2007).
Three polyphagous Neotropical Liriomyza species (Liriomyza huidobrensis (Blanchard), Liriomyza sativae Blanchard, and Liriomyza trifolii (Burgess)) have become established around the world (Murphy and La Salle, Reference Murphy and LaSalle1999; Scheffer and Lewis, Reference Scheffer and Lewis2001, Reference Scheffer and Lewis2005, Reference Scheffer and Lewis2006; Weintraub et al., Reference Weintraub, Scheffer, Visser, Valladares, Correa, Shepard, Rauf, Murphy, Mujica, MacVean, Kroschel, Kishinevsky, Joshi, Johansen, Hallett, Çivelek, Chen and Metzler2017), largely through the movement of infested plant material along trade routes (Minkenberg, Reference Minkenberg1988). Despite strict surveillance and quarantine programs, these three species are now established in mainland Australia (Xu et al., Reference Xu, Ridland, Umina, Gill, Ross, Pirtle and Hoffmann2021a). Liriomyza sativae was detected for the first time in the islands of the Torres Strait in 2008 (Blacket et al., Reference Blacket, Rice, Semeraro and Malipatil2015), and then on the Australian mainland at Seisia in 2015 (IPPC 2017). This species has not been detected outside of Cape York for the last 7 years. Liriomyza huidobrensis was first confirmed at several sites in western Sydney and then in southern Queensland in 2020 (IPPC 2021a; Mulholland et al., Reference Mulholland, Gopurenko, Mirringto, Löcker, Gillespie, Rossiter and Anderson2022). Liriomyza trifolii was first detected in 2021 in Kununurra (northern Western Australia), Bamaga (Far North Queensland), and the Torres Strait (IPPC 2021b).
Worldwide, farmers have routinely relied on synthetic chemical pesticides to control leafmining flies. However, the extensive use of pesticides has dramatically reduced their effectiveness due to the rapid evolution of resistance within some species and the negative impacts of these chemicals on natural enemies (Reitz et al., Reference Reitz, Gao, Lei and Tradan2013). Natural enemies, in particular parasitoid wasps, have now been recognized as effective bio-control agents that can suppress outbreaks of leafmining pests; indigenous parasitoid wasps can quickly suppress recent invasions of Liriomyza flies in pesticide-free areas (Murphy and La Salle, Reference Murphy and LaSalle1999; Liu et al., Reference Liu, Kang, Heinz and Trumble2009). In addition, some countries have introduced exotic parasitoid wasps to control Liriomyza spp. with encouraging results. For example, Chrysocharis oscinidis Ashmead and Banacuniculus utilis (Beardsley) (Hymenoptera: Figitidae: Eucoilinae) were successfully introduced to Hawaii, Guam, and Tonga for the management of L. sativae and L. trifolii (Johnson, Reference Johnson1993), while Japan imported Diglyphus isaea (Walker) (Hymenoptera: Eulophidae) and Dacnusa sibirica Telenga (Hymenoptera: Braconidae) from Europe for augmentative biological control of L. trifolii in greenhouses (Mitsunaga and Yano, Reference Mitsunaga and Yano2004).
Australia already has a suite of endemic and introduced parasitoids that parasitize a range of adventive and endemic agromyzid species, which would likely contribute to the regulation of exotic Liriomyza pests (Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020). It has been demonstrated that agromyzid species colonizing weeds and non-crop plants serve as useful reservoirs to support parasitoid wasps, potentially providing parasitoids for the biological control of invasive Liriomyza spp. (Lardner, Reference Lardner1991; Bjorksten et al., Reference Bjorksten, Robinson and La Salle2005; Lambkin et al., Reference Lambkin, Fayed, Manchester, La Salle, Scheffer and Yeates2008; Wood et al., Reference Wood, Siekmann, Stephens, DeGraaf, La Salle and Glatz2010; Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020). However, numerous challenges impede the uptake of augmentative or conservation biological control programs including indiscriminate use of pesticides, lack of parasitoid reservoirs in intensive cropping systems, and the high cost of mass-rearing for augmentative biological control.
Accurate species identification of parasitoids underpins all these issues. Traditional morphological identification of parasitoids relies on taxonomic keys, which can be difficult to use by non-specialist researchers. Most published studies surveying parasitoids of agromyzids (e.g. Asadi et al., Reference Asadi, Talebi, Fathipour, Moharramipour and Rakhshani2006; Lambkin et al., Reference Lambkin, Fayed, Manchester, La Salle, Scheffer and Yeates2008; Mujica and Kroschel, Reference Mujica and Kroschel2011) have relied on morphological identification. However, the dearth of taxonomic expertize worldwide remains a major limiting factor for the authoritative morphological identification of specimens. High-resolution morphological information can now be collected more easily by non-specialists using techniques such as scanning electron microscopy (SEM) and multi-focus imaging, but still require validation from experts.
DNA barcodes are increasingly being used to supplement morphological studies in the identification of parasitoid wasps (Powell et al., Reference Powell, Caleca, Sinno, van Staden, van Noort, Rhode, Allsopp and van Asch2019). Some taxonomic studies now combine molecular data, morphological data and high-quality imaging in assessing parasitoids of leafminers such as eulophids attacking agromyzids (e.g. Perry and Heraty, Reference Perry and Heraty2019, Reference Perry and Heraty2021). However, morphological misidentifications and incorrect DNA barcodes on public databases hinder research assessing the impact of parasitoids (Lue et al., Reference Lue, Buffington, Scheffer, Lewis, Elliott, Lindsey, Driskell, Jandova, Kimura, Carton and Kula2021, Reference Lue, Abram, Hrcek, Buffington and Staniczenko2022). This is an important issue, because validated DNA barcodes can be used by researchers with little taxonomic experience (Darling and Blum, Reference Darling and Blum2007) and can also be applied to immature life stages and cryptic species (Waugh, Reference Waugh2007). Nevertheless, accurate DNA barcodes depend on the use of specimens that have been authoritatively identified (Lue et al., Reference Lue, Buffington, Scheffer, Lewis, Elliott, Lindsey, Driskell, Jandova, Kimura, Carton and Kula2021, Reference Lue, Abram, Hrcek, Buffington and Staniczenko2022).
As well as assisting in identifying parasitoid species, DNA technology can be used to screen for the presence of bacterial endosymbionts within parasitoid specimens. Endosymbionts like Wolbachia are intracellular bacteria that are widespread in arthropod species, including parasitoid wasps (Floate et al., Reference Floate, Kyei-Poku and Coghlin2006; Klopfstein et al., Reference Klopfstein, van Der Schyff, Tierney and Austin2018). Wolbachia is often associated with host reproductive effects that include male-killing, feminization, parthenogenesis, and cytoplasmic incompatibility (CI) (Sinkins et al., Reference Sinkins, Curtis, O'Neill, O'Neill, Hoffman and Werren1997). Other endosymbionts such as Cardinium and Rickettsia are also common in insects and can affect traits such as reproduction (Montenegro et al., Reference Montenegro, Solferini, Klaczko and Hurst2005; Hagimori et al., Reference Hagimori, Abe, Date and Miura2006). To date, endosymbiont surveys of parasitoids of agromyzids are limited (Tagami et al., Reference Tagami, Doi, Sugiyama, Tatara and Saito2006), despite these endosymbionts having the potential to be used to generate strains of parasitoids with useful characteristics for future release. Moreover, endosymbionts can affect patterns of mtDNA variation (e.g. Wolbachia induced CI decreases mtDNA polymorphism as the Wolbachia and its associated mtDNA variant spreads in a population) and influence within-species variation in clades identified from mtDNA markers (Hale and Hoffmann, Reference Hale and Hoffmann1990).
Prior studies have found 27 genera of parasitoids of agromyzids in Australia, and some are likely to be important in controlling the invasive polyphagous Liriomyza species (Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020). Therefore, based on this background, our study mainly focuses on parasitoid wasp species reared from common agromyzids in Australia, aiming to determine which species are present and potentially helpful in suppressing the exotic Liriomyza pests. We compared DNA barcodes with sequences on public databases and verified morphological identifications where possible with SEM images. For species without DNA barcodes, we provide DNA information alongside morphological descriptions. Furthermore, we assessed endosymbiont infections in parasitoid wasps that might potentially be exploited for future augmentative biocontrol.
Materials and methods
Insects
We reared parasitoid wasps from three adventive agromyzid species: Liriomyza brassicae (Riley), Phytomyza plantaginis Goureau and Phytomyza syngenesiae (Hardy), and two native agromyzid species: Liriomyza chenopodii (Watt) and Phytoliriomyza praecellens Spencer. These agromyzids are likely to be reservoirs for parasitoids to attack invasive Liriomyza pests (Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020).
Mined plant leaves were primarily collected from locations in Melbourne (Victoria, Australia) and Bangalow (New South Wales, Australia). Samples reared from Liriomyza huidobrensis were received from Wyreema (Queensland, Australia). Table 1 gives detailed information on sampling. We also received samples of Diglyphus isaea (ex L. bryoniae) from a laboratory colony (Koppert BV) in The Netherlands and Hemiptarsenus varicornis (Girault) (ex L. trifolii) from Fiji. This enabled us to compare overseas accessions of these two widely distributed parasitoid species with Australian specimens, both in terms of COI haplotype and endosymbiont status. Leaf samples were first cleared of other insects and residues, then covered with paper towels and placed into individual Ziploc® bags (SC Johnson, Australia). Paper toweling was changed frequently to reduce the moisture content within the Ziploc® bags. We checked the bags regularly, and when adult flies and parasitoid wasps emerged within each bag, we removed and separated them (avoiding teneral insects, which can be difficult to identify). Flies and parasitoids were preserved in absolute ethanol and stored at −20°C for DNA extractions, while some parasitoid wasps were placed in 70% ethanol and stored at 4°C for SEM imaging. Different individuals of the same wasp species were processed in two ways. Approach 1, where whole bodies were used to extract DNA and detect symbiotic bacteria. Approach 2, where body parts (e.g. legs) were used for DNA extraction and SEM was undertaken on key body components to assist in species identification. The identifications of the agromyzid fly species were confirmed with the DNA barcodes we published previously (Coquilleau et al., Reference Coquilleau, Xu, Ridland, Umina and Hoffmann2021; Xu et al., Reference Xu, Coquilleau, Ridland, Umina, Yang and Hoffmann2021b). Voucher specimens were deposited in the Victorian Agricultural Insect Collection, AgriBio.
Table 1. The COI haplotypes and endosymbionts detected in parasitoid wasps characterized in this study
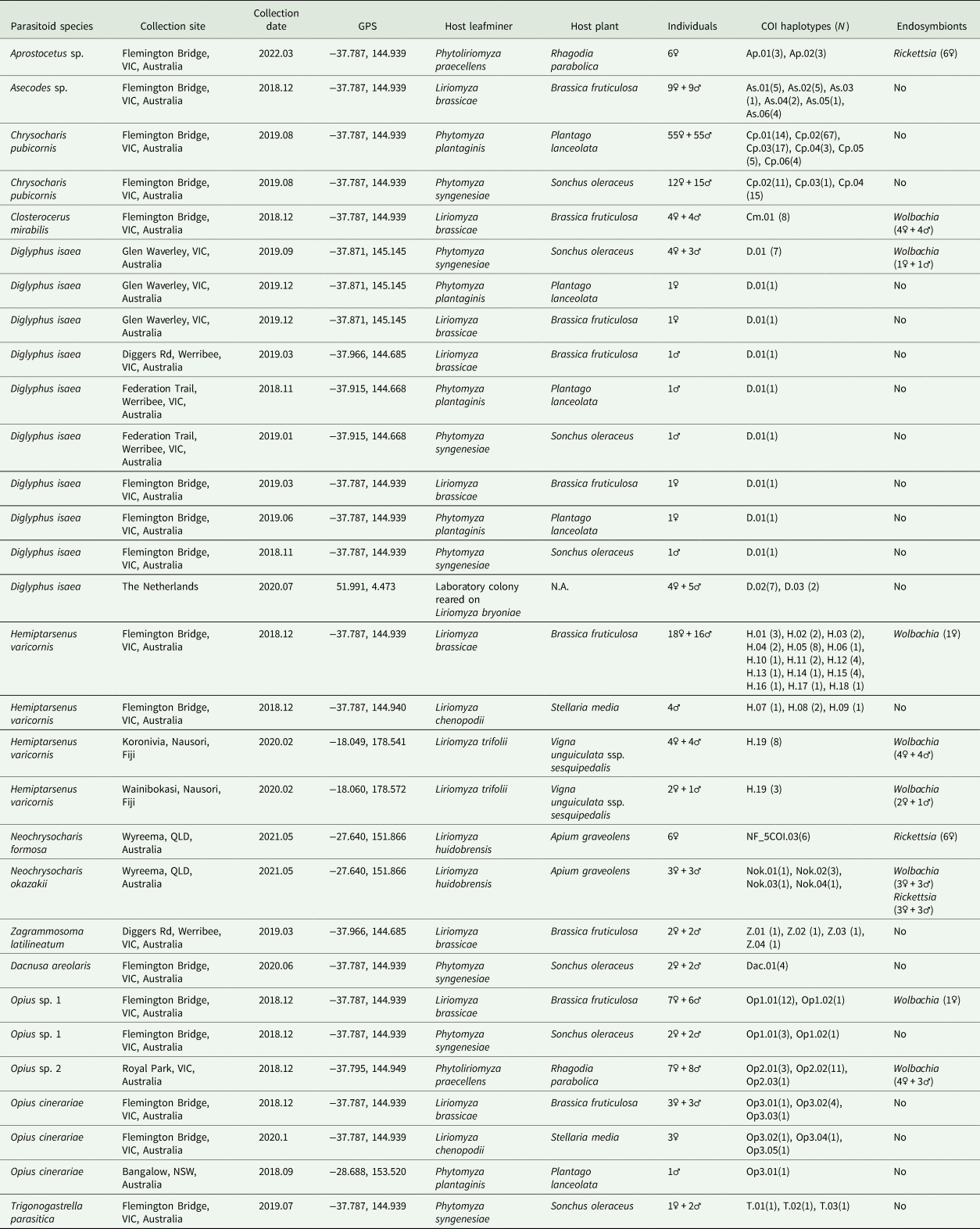
VIC, Victoria; QLD, Queensland; NSW, New South Wales.
DNA extraction, amplification, and sequencing
We extracted genomic DNA using a Chelex (Bio-Rad Laboratories) DNA extraction method. For approach 1, DNA was extracted from whole parasitoid bodies, which involved crushing the body with two glass beads (3 mm) in 100 μl of 5% Chelex solution. For approach 2, we extracted DNA from the legs of wasps in order to preserve the remaining body parts for SEM imaging. For particularly tiny wasps (<1.5 mm in length), such as Asecodes sp. (Hymenoptera: Eulophidae) and Closterocerus mirabilis Edwards & La Salle (Hymenoptera: Eulophidae), we failed to successfully extract sufficient DNA from legs, so in these instances, we used the head & legs or the gaster and legs to extract DNA (in a total volume of 70 μl of 5% Chelex solution). In all cases, the Chelex mixture was incubated with 3 μl proteinase K (20 mg ml−1, Roche Diagnostics) at 65°C for 1 h. The homogenates were then boiled at 95°C for 10 min to inactivate the proteinase K and these samples were used as templates for PCR.
The DNA barcodes we used in this study focused on the 5′ region of the cytochrome c oxidase subunit I (COI) gene. Additional sequences were obtained by sequencing the genes for some species (e.g., nuclear ribosomal internal transcribed spacer 1 (ITS1) sequences for D. isaea, 28S sequences for Neochrysocharis okazakii Kamijo (Hymenoptera: Eulophidae), and 3′ region COI sequences for Asecodes sp., N. okazakii and Zagrammosoma latilineatum Ubaidillah (Hymenoptera: Eulophidae)). For detailed information on primers used for DNA barcoding see table S1. It is noteworthy that primers LepF1/LepR1 used for Cl. mirabilis failed to sequence the COI gene because the endosymbiont Wolbachia was sequenced instead of the target. In this case, we undertook further DNA extractions following the methods described above, except we only used the head and legs due to the low Wolbachia densities in these organs (Narita et al., Reference Narita, Nomura and Kageyama2007; Frentiu et al., Reference Frentiu, Zakir, Walker, Popovici, Pyke, van den Hurk, McGraw and O'Neill2014; Amuzu and McGraw, Reference Amuzu and McGraw2016).
PCRs for DNA barcoding involved 2 μl DNA template, 3 μl 10 × ThermoPol® Reaction Buffer (New England BioLabs: B9004S), 2.4 μl dNTPs (2.5 mM), 1.5 μl of both forward and reverse primers (10 μM), and 0.3 μl BSA (New England BioLabs, B9000S), 0.2 μl Taq polymerase (New England BioLabs: M0267X) and ddH2O to create a final 30 μl reaction volume. All PCRs included a sterile water sample (without genomic DNA) to confirm there was no DNA contamination. PCR products were directly sequenced in both directions using the primers detailed in table S1 at Macrogen (Seoul, Korea).
We determined the infection status of parasitoid wasps with three common endosymbionts (Wolbachia, Cardinium, and Rickettsia) using PCR reactions (and primers) described in Tagami et al. (Reference Tagami, Doi, Sugiyama, Tatara and Saito2006). In brief, wsp sequence was selected to identify Wolbachia status and specific 16S rDNA sequences were selected to assess the presence of Cardinium/Rickettsia.
Sequence analysis
DNA sequences of parasitoid wasps were aligned and manually edited using Geneious 9.1.8 (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran and Thierer2012). Sequence similarities were searched first through BLAST (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990; Ratnasingham and Hebert, Reference Ratnasingham and Hebert2007) and when matches were identified, we checked whether specimens had been identified. All available sequences were then downloaded from the BOLD System (Ratnasingham and Hebert, Reference Ratnasingham and Hebert2007) and the NCBI GenBank database (Benson et al., Reference Benson, Cavanaugh, Clark, Karsch-Mizrachi, Ostell, Pruitt and Sayers2018) and combined for further analyses. Polymorphism levels (haplotype and nucleotide diversity) were calculated using DnaSP version 6 (Rozas et al., Reference Rozas, Ferrer-Mata, Sánchez-DelBarrio, Guirao-Rico, Librado, Ramos-Onsins and Sánchez-Gracia2017). In this study, haplotype divergence within a specific species includes any sequence difference detected even if it involves a difference of only one nucleotide between the sequences. We excluded the duplicate haplotypes in the public dataset and the genetic cluster analysis. All other different haplotypes are included. A Neighbor-Joining tree (Kimura-2 parameter model) was generated with 1000 bootstrap replications using MEGA X (Kumar et al., Reference Kumar, Stecher, Li, Knyaz and Tamura2018). Pairwise genetic distances were calculated to assess the genetic similarity of sequences.
In those instances where the endosymbiont Wolbachia was detected, we allocated sequences to Wolbachia supergroups. To do this, wsp sequences from previously confirmed data (Baldo et al., Reference Baldo, Dunning Hotopp, Jolley, Bordenstein, Biber, Choudhury, Hayashi, Maiden, Tettelin and Werren2006) were obtained to construct a phylogenetic tree and allocate Wolbachia supergroups.
Morphological identification and scanning electron micrographs
Individual wasps were identified using Lucid keys (Reina and La Salle, Reference Reina and La Salle2003; Fisher et al., Reference Fisher, Ubaidillah, Reina and La Salle2005) and multiple published papers specific to each genus or species: Aprostocetus sp. (Bouček, Reference Bouček1988; Yang et al., Reference Yang, Lin, Wu, Fisher, Saimanee, Sangtongpraow, Zhu, Chiu and La Salle2014), Asecodes sp. (Bouček, Reference Bouček1988; Hansson, Reference Hansson1994, Reference Hansson1996), Chrysocharis pubicornis (Hansson, Reference Hansson1985, Reference Hansson1987; Bouček, Reference Bouček1988; Ikeda, Reference Ikeda1995, Reference Ikeda1996), Closterocerus mirabilis (Edwards and La Salle, Reference Edwards and La Salle2004), D. isaea (Hansson and Navone, Reference Hansson and Navone2017); Hemiptarsenus (Bouček, Reference Bouček1988; Fisher et al., Reference Fisher, Ubaidillah, Reina and La Salle2005), Neochrysocharis formosa (Westwood) (Hansson, Reference Hansson1990; Fisher et al., Reference Fisher, Ubaidillah, Reina and La Salle2005), N. okazakii (Kamijo, Reference Kamijo1978), Z. latilineatum (Ubaidillah et al., Reference Ubaidillah, LaSalle and Rauf2000; Perry and Heraty, Reference Perry and Heraty2021); Trigonogastrella parasitica Girault (Bouček, Reference Bouček1988), Dacnusa areolaris (Nees) (Wharton and Austin, Reference Wharton and Austin1991), Opius cinerariae Fischer, Opius atricornis Fischer and Opius oleracei Fischer (Belokobylskij et al., Reference Belokobylskij, Wharton and La Salle2004). For detailed morphological information, see Supplementary Information (figs S1–S10).
Scanning electron micrograph (SEM) images were produced on an FEI Teneo Volumescope instrument (ThermoFisher Scientific, USA) at an operating voltage of 10 kV. Wasp specimens were dissected and fixed on the SEM specimen mount by double-sided carbon tape, followed by air drying for four hours. A 5 nm gold coating was applied to the samples using an Emitech K575x sputter coater (Quorum Technologies, Canada) before taking SEM images.
Results
Wasp identifications
Overall, we characterized 307 parasitoid individuals from 10 geographic locations (seven from Australia, two from Fiji, and a laboratory colony from The Netherlands). We found 14 species based on morphological identifications and DNA barcoding. Among these, eight species could be linked to DNA barcodes in public databases: C. pubicornis, D. isaea, Dac. areolaris, H. varicornis, N. formosa, N. okazakii, O. cinerariae, and Z. latilineatum. We compared our COI barcodes with sequences from databases to examine levels of genetic variation within taxa. For species without DNA barcodes in either BOLD or GenBank or where there was substantial sequence divergence (>5%), we provide additional morphological analyses (figs S1–S10). DNA sequences generated in this study were deposited in GenBank under accession numbers (table S2).
Chrysocharis pubicornis
All specimens were collected from the Flemington Bridge location, in Melbourne. Among these, 110 individuals were collected from mined leaves of Plantago lanceolata (host flies were P. plantaginis) and 27 individuals were collected from mined leaves of Sonchus oleraceus (host flies were P. syngenesiae). The 5′ region COI sequences of C. pubicornis revealed six haplotypes (Cp.01–Cp.06) were present in our study and the dominant haplotype was Cp.02 (representing 56.9% of sequences). Genetic divergence of C. pubicornis haplotypes in this study varied from 0.2 to 6.3% based on 409 bp COI sequences (table S3). However, we were unable to detect morphological differences between these haplotypes using taxonomic keys and SEM imaging.
A phylogenetic tree was generated that included specimens from our study and sequences of C. pubicornis from public databases (fig. 1). The genetically related species Chrysocharis pallipes (Nees) (Hymenoptera: Eulophidae) was used as an outgroup based on recent studies of the phylogenetic relationship of Hymenoptera (Derocles et al., Reference Derocles, Evans, Nichols, Evans and Lunt2015). Phylogenetic analyses suggested at least five major COI clades in C. pubicornis: C. pubicornis-A–E. This and the presence of substantial sequence divergence suggests the possibility of cryptic species within this taxon. Chrysocharis pubicornis collected in Australia are separated into two clades. Haplotypes Cp.01 and Cp.02 are within clade C. pubicornis-A, together with samples from Canada, the UK, and Norway. Genetic variation in clade C. pubicornis-A ranges from 0.2 to 1.4%. C. pubicornis-B is nearby to clade C. pubicornis-A and includes samples from Germany and Canada, with a genetic variation of 1.4%. Australian haplotypes Cp.03 to Cp.06 are within clade C. pubicornis-C alone, with genetic variation ranging from 0.2 to 0.4%. C. pubicornis-D includes samples from Belarus, Germany, and Japan, with genetic variation ranging from 0.2 to 5.6%. C. pubicornis-E includes samples from the UK, Canada and Norway, with genetic variation ranging from 0.2 to 2.2%. The genetic distances across all C. pubicornis samples ranged from 0.2 to 9.0% (table 2). The smallest genetic distances between C. pubicornis samples and outgroup species Chrysocharis pallipes was 9.5% (table S3)
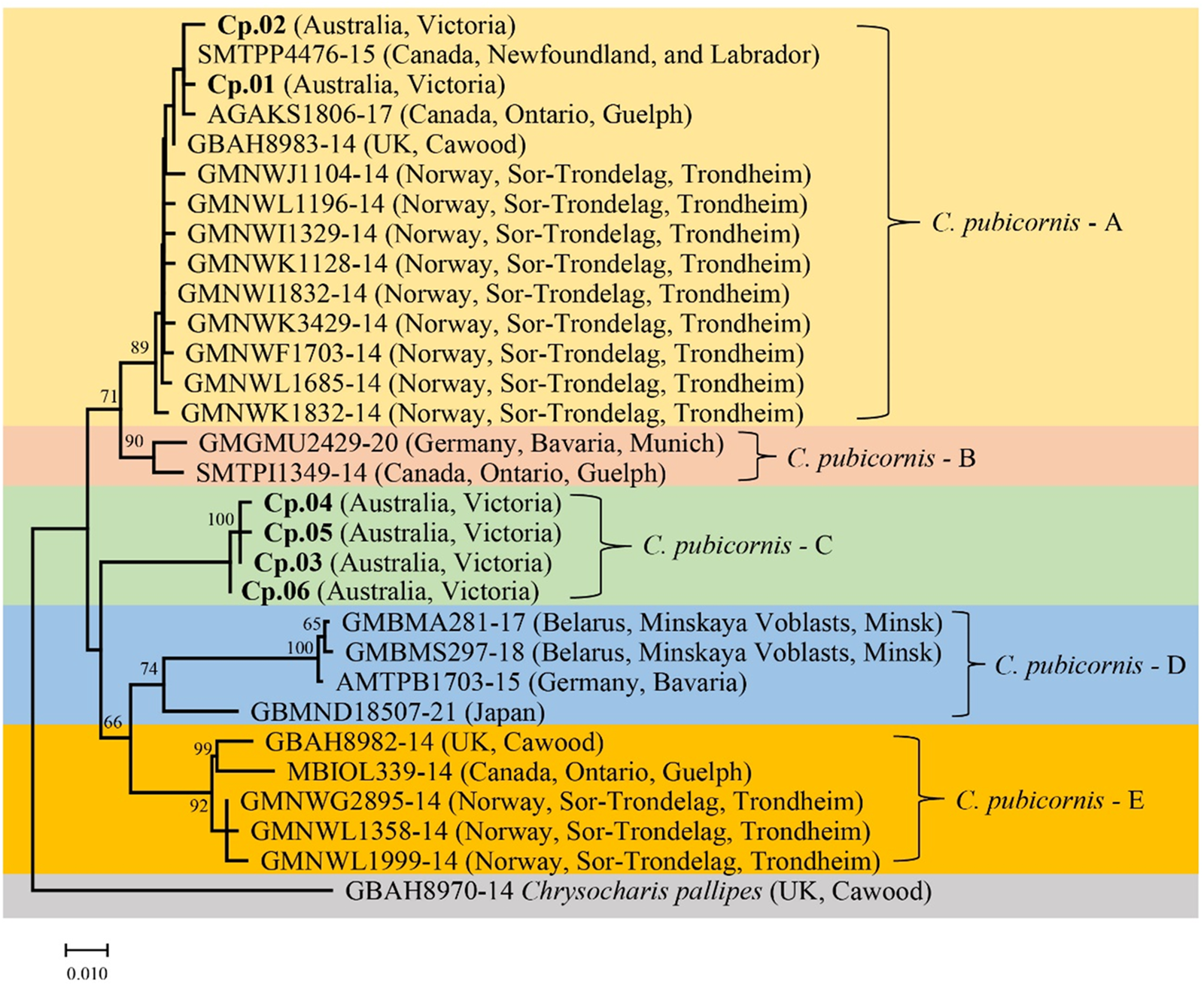
Figure 1. COI phylogenetic tree of Chrysocharis pubicornis was generated using the Neighbor-Joining method (1000 bootstrap replications, Kimura-2 parameter model) based on 409 bp sequence data. The scale bar indicates nucleotide substitutions per site. Haplotypes in this study are highlighted in bold, and the remainder of the sequences are from the BOLD database. Chrysocharis pallipes is the outgroup species.
Table 2. Uncorrected pairwise distances among Chrysocharis pubicornis clades based on 409 bp COI sequence data

Diglyphus isaea
In total, 15 D. isaea individuals were collected from three host species (L. brassicae, P. plantaginis, and P. syngenesiae) (table 1) in Melbourne sites, and only one COI haplotype (D.01) was found. We detected two COI haplotypes (D.02 and D.03) in nine D. isaea individuals obtained from the Netherlands. Sha et al. (Reference Sha, Zhu, Murphy, La Salle and Huang2006) suggested that D. isaea in China is very likely a complex of cryptic species because five main clades were identified based on COI sequences and this was further supported by ITS1 sequences (Sha et al., Reference Sha, Zhu, Murphy and Huang2007). In this study, we reconstructed the COI phylogenetic tree including our sequences of D. isaea and those from China (fig. 2). We found D. isaea from Australia and the Netherlands clustered together in Group I, the largest clade, which also included D. isaea from northern and southern populations in China.

Figure 2. COI phylogenetic tree of Diglyphus isaea was generated using the Neighbor-Joining method (1000 bootstrap replications, Kimura-2 parameter model) based on 745 bp sequence data. The scale bar indicates nucleotide substitutions per site. Haplotypes in this study are highlighted in bold. The remainder of the sequences of D. isaea and the outgroup D. bimaculatus are from China and taken from Sha et al. (Reference Sha, Zhu, Murphy, La Salle and Huang2006).
Hemiptarsenus varicornis
Forty-nine H. varicornis individuals were collected from three host species (L. brassicae, L. chenopodii and L. trifolii) (table 1) with 18 COI haplotypes (H.01–H.18) identified in Victoria and one COI haplotype (H.19) found from Fiji. There are 77 H. varicornis COI sequences in the BOLD database. Alignment yielded a 375 bp fragment, allowing us to incorporate these BOLD sequences (table S4) into our analysis, which revealed a further 20 haplotypes. The phylogenetic tree demonstrated three major clades within this species: H. varicornis-A, H. varicornis-B, and H. varicornis-C (fig. 3). Clade H. varicornis-A represents all H. varicornis from Australia; clade H. varicornis-B is a single specimen from Fiji and this clade is nearby to Clade H. varicornis-C, which includes specimens from Pakistan, Malaysia, and Vietnam. Notably, the COI sequence of H. varicornis (AB721362) in GenBank is nearly identical to N. okazakii (AB721363) (with only three base pair differences) (Nakamura et al., Reference Nakamura, Masuda, Mochizuki, Konishi, Tokumaru, Ueno and Yamaguchi2013). We suspect that these two sequences are both incorrect (neither H. varicornis nor N. okazakii). The uncorrected pairwise distances within clade H. varicornis-A + B (haplotype H.01–H.34) was 0.2–3.2% while the genetic distances within clade H. varicornis-C (haplotype H.35–H.38) were 0.2–0.8% (table S5). There is a clear genetic divergence between Australian H. varicornis and other locations with the largest divergence being 3.5% (haplotype H.35-Vietnam and haplotype H.24-WA, Australia) (table S5).

Figure 3. COI phylogenetic tree of Hemiptarsenus varicornis was generated using the Neighbor-Joining method (1000 bootstrap replications, Kimura-2 parameter model) based on 375 bp sequence data. The scale bar indicates nucleotide substitutions per site. Information about the haplotypes used in this study can be found in table S4. Pnigalio maculipes is the outgroup species.
Opius spp.
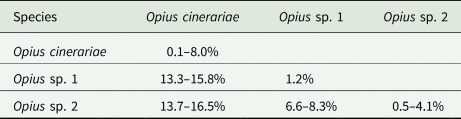
We detected three Opius species with one species identified to species level as O. cinerariae and the other two putative species based on morphology and sequence divergence identified to genus level (Opius sp. 1 and Opius sp. 2). We found two COI haplotypes (Op1.01 and Op1.02) in Opius sp. 1, which were reared from two host flies (L. brassicae and P. syngenesiae) (table 1). The dominant haplotype was Op1.01, which accounted for 88% of the samples. For Opius sp. 2, we found three haplotypes (Op2.01, Op2.02, and Op2.03), all from P. praecellens. Op2.02 was the dominant haplotype, accounting for 73% of Opius sp. 2 samples. For O. cinerariae, five COI haplotypes (Op3.01–Op3.05) were detected from three host flies (L. brassicae, L. chenopodii, and P. plantaginis) (table 1). Among these, Op3.01–Op3.03 were reared from L. brassicae and P. plantaginis, while Op3.04–Op3.05 were only reared from L. chenopodii. A phylogenetic tree based on 577 bp COI sequence data was constructed and showed clear species boundaries among the three Opius species (fig. 4). Opius sp. 1 is genetically closer to Opius sp. 2 (uncorrected pairwise distances ranging from 6.6–8.3%) than O. cinerariae, (fig. 4, table 3, table S6). Furthermore, genetic divergence is evident within the O. cinerariae clade. The uncorrected pairwise distances of O. cinerariae from Australia (in this study) and New Zealand (NZHYM868–11 and NZHYM870–11) range from 6.9 to 8.0%, pointing to the possibility of cryptic species.
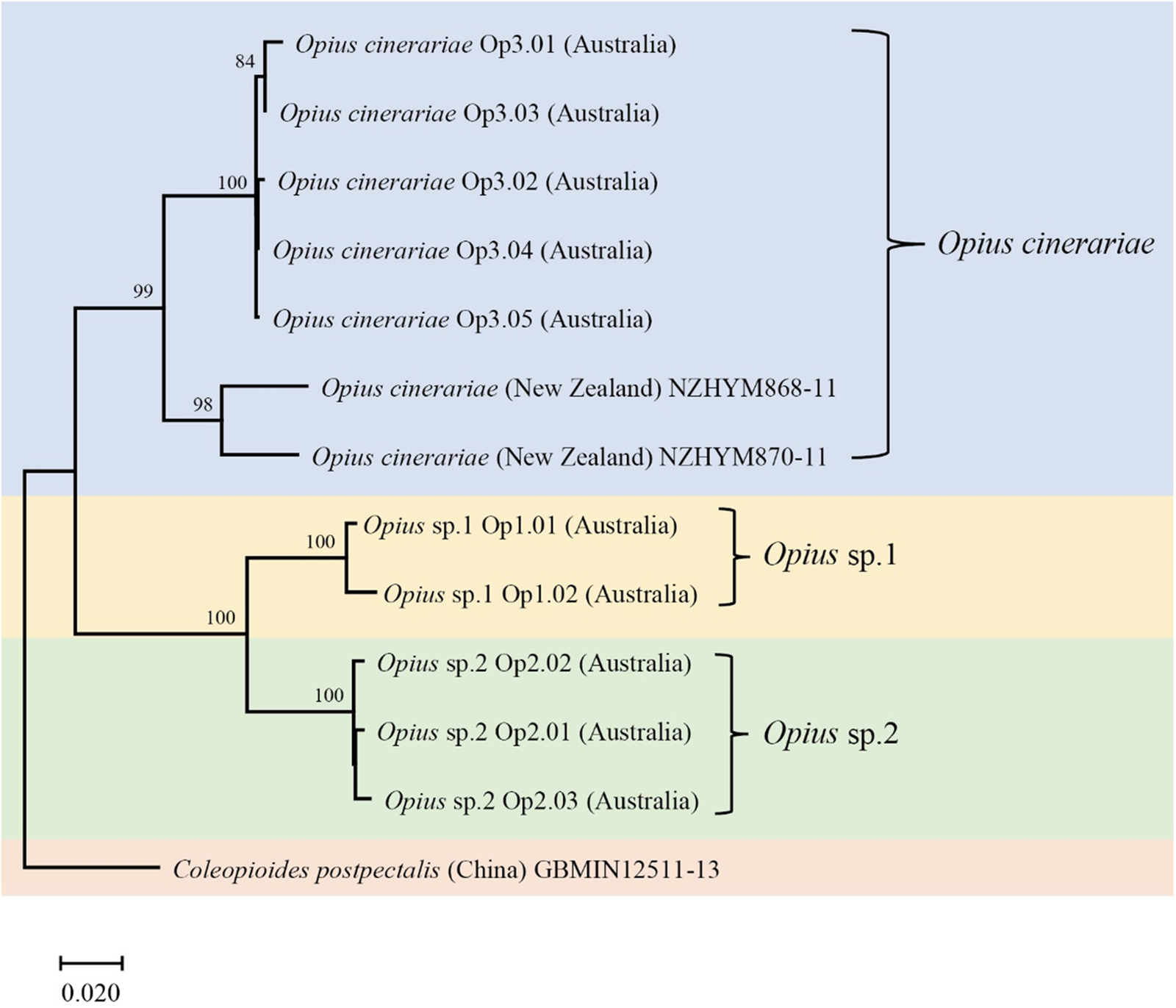
Figure 4. The COI phylogenetic tree of Opius spp. was generated using the Neighbor-Joining method (1000 bootstrap replications, Kimura-2 parameter model) based on 577 bp sequences. The scale bar indicates nucleotide substitutions per site. Coleopioides postpectalis was the outgroup species (Li et al., Reference Li, van Achterberg and Tan2013).
Table 3. Uncorrected pairwise distances among Opius species based on 577 bp COI sequence data
Other parasitoid species
There are two species (Dac. areolaris, N. okazakii) with DNA barcodes on BOLD and four species (Aprostocetus sp., Asecodes sp., Cl. mirabilis, and T. parasitica) without DNA barcodes. In this study, we provide both morphological and COI information for these six species (table 1, figs S1–S3, S5–S7). For Dac. areolaris, we only found a single COI haplotype (Dac.01) and sequences were 99.8% similar to specimens from Germany (BOLD: GBMIX500–14). For N. okazakii, 28S sequences were also obtained and indicated our samples from Queensland are 100% identical to N. okazakii (NCBI: AB526861) from Japan (Adachi-Hagimori et al., Reference Adachi-Hagimori, Miura and Abe2011). However, the 3′ COI gene region results suggest our N. okazakii from Queensland are not the same species as N. okazakii (NCBI: AB721363) based on sequences provided by Nakamura et al. (Reference Nakamura, Masuda, Mochizuki, Konishi, Tokumaru, Ueno and Yamaguchi2013). The pairwise distances between our N. okazakii sequences and these N. okazakii (NCBI: AB721363) were in the range of 15.4–16.1%. However, we consider the N. okazakii sequences provided by Nakamura et al. (Reference Nakamura, Masuda, Mochizuki, Konishi, Tokumaru, Ueno and Yamaguchi2013) problematic given that their COI sequences for N. okazakii (NCBI: AB721363.1) are identical to H. varicornis (NCBI: AB721362). Our morphological identification also supported our conclusion that our specimen is N. okazakii. (fig. S5). We provide the first COI barcodes for Aprostocetus sp., Asecodes sp., Cl. mirabilis, and T. parasitica. Among these, we found two haplotypes for Aprostocetus sp., six haplotypes for Asecodes sp., one haplotype for Cl. mirabilis, and three haplotypes for T. parasitica (table 1).
Prior studies already provide detailed molecular and morphological data on N. formosa and Z. latilineatum (Perry and Heraty, Reference Perry and Heraty2021; Xu et al., Reference Xu, Hoffmann, Umina, Coquilleau, Gill and Ridland2022), so we compared our COI data with these sequences. For N. formosa, Xu et al. (Reference Xu, Hoffmann, Umina, Coquilleau, Gill and Ridland2022) found two 5′ COI haplotypes (NCBI: OK076720, NF_5COI.01 and OK076721, NF_5COI.02) from Victorian collections. In this study, we only found one 5′ COI haplotype (NF_5COI.03) for N. formosa from Queensland, which is a single base pair different from NF_5COI.02. For Z. latilineatum, Perry and Heraty (Reference Perry and Heraty2021) only provided 3′ COI sequences (NCBI: MK753233, Australia), which were 0.5–2.0% similar to the Z. latilineatum sequences we generated in this study.
Endosymbiont detections
In the Australian samples, we found H. varicornis (1 positive/38 total), D. isaea (2/24), Cl. mirabilis (8/8), Opius sp. 1 (1/17), and Opius sp. 2 (7/14) infected with Wolbachia, but often at low frequency. In contrast, Aprostocetus sp. and N. formosa samples were uniformly infected with Rickettsia sp. (table 1). Additionally, we found N. okazakii infected with two endosymbionts (Wolbachia and Rickettsia sp.) simultaneously. Based on these findings, we constructed a phylogenetic tree using Wolbachia wsp sequences (fig. 5). The Wolbachia wsp sequences of Cl. mirabilis, Opius sp. 1, and D. isaea were identical and belong to Wolbachia Supergroup A. This wsp sequence is identical to the wsp sequence of L. sativae (wLsatC) from Vietnam (Xu et al., Reference Xu, Ridland, Umina, Gill, Ross, Pirtle and Hoffmann2021a). The Wolbachia wsp sequences of H. varicornis and Opius sp. 2 belong to Wolbachia Supergroup B. There is only a single base pair difference between the Wolbachia wsp sequences of H. varicornis from Fiji (this study where all 11 specimens were infected) and Japan (NCBI: AB231470.1/AB231471.1) (Tagami et al., Reference Tagami, Doi, Sugiyama, Tatara and Saito2006). Additionally, the Wolbachia wsp sequence of H. varicornis from Fiji is identical to the wsp sequence of L. bryoniae from Japan (wLbryB) (Tagami et al., Reference Tagami, Doi, Sugiyama, Tatara and Saito2006; Xu et al., Reference Xu, Ridland, Umina, Gill, Ross, Pirtle and Hoffmann2021a). The Wolbachia wsp sequence of H. varicornis from Australia (this study) is three base pairs different to the wsp sequence of L. bryoniae (wLbryA) from the Netherlands (Xu et al., Reference Xu, Ridland, Umina, Gill, Ross, Pirtle and Hoffmann2021a). The Wolbachia wsp sequence of Opius sp. 2 is previously undescribed and we did not find similar sequences in any other host.
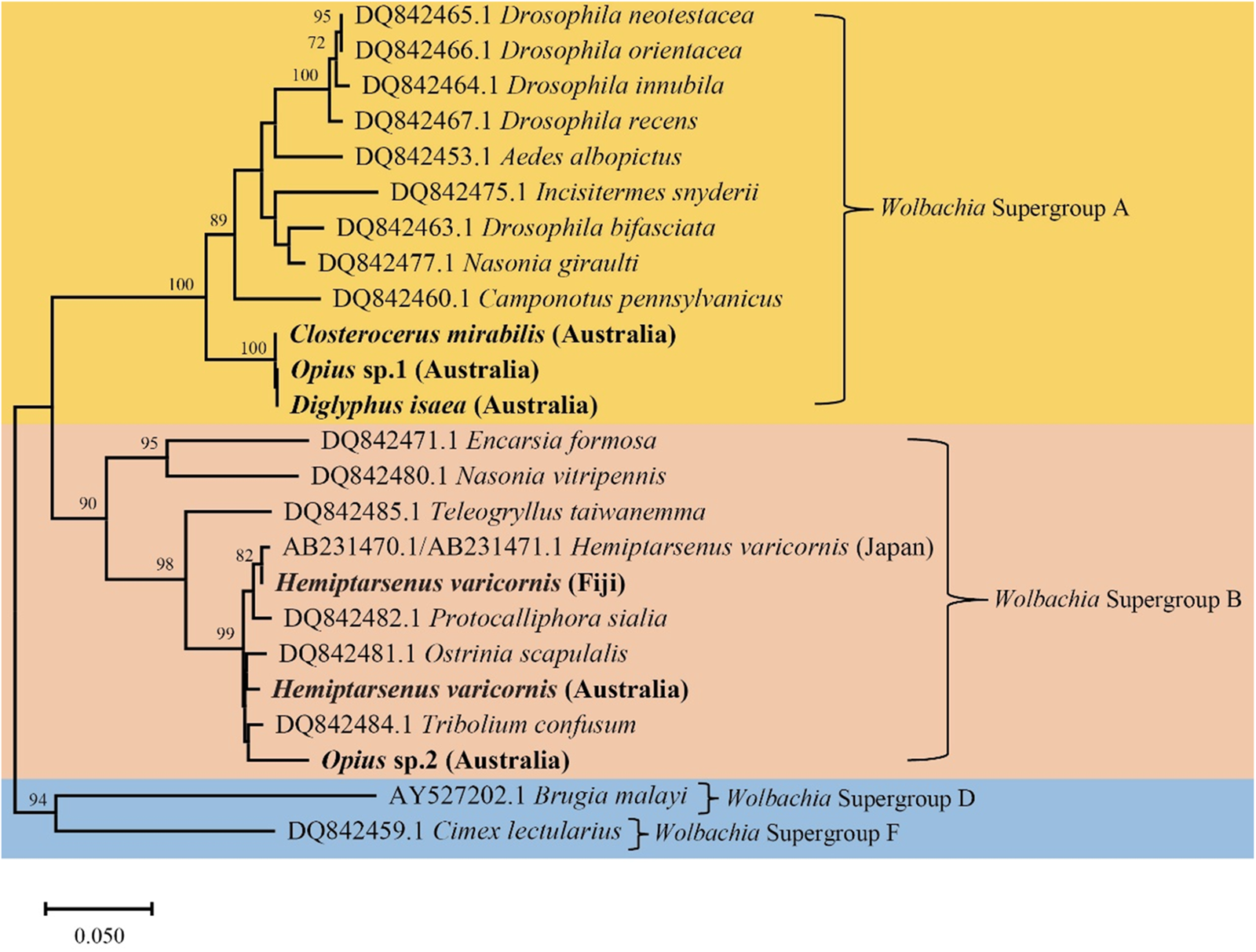
Figure 5. Phylogenetic tree of Wolbachia wsp sequences in different insect hosts generated using the Neighbor-Joining method (1000 bootstrap replications, Kimura-2 parameter model) based on 382 bp sequence data. The scale bar indicates nucleotide substitutions per site. Species in this study are highlighted in bold and the remainder of the sequences are from Baldo et al. (Reference Baldo, Dunning Hotopp, Jolley, Bordenstein, Biber, Choudhury, Hayashi, Maiden, Tettelin and Werren2006).
Both N. formosa and N. okazakii were infected with the same Rickettsia sp. based on 253 bp COI sequence data, which is identical to N. formosa previously screened from Australia, Japan, and China (Xu et al., Reference Xu, Hoffmann, Umina, Coquilleau, Gill and Ridland2022). The Rickettsia sp. sequence of Aprostocetus sp. is only two base pairs different to N. formosa and N. okazakii sequences generated in this study. Moreover, N. okazakii is infected with Wolbachia, with the wsp sequence identical to L. brassicae (NCBI: MW047082.1) (Xu et al., Reference Xu, Ridland, Umina, Gill, Ross, Pirtle and Hoffmann2021a).
Discussion
Biological control has become important in managing L. huidobrensis, L. sativae and L. trifolii, largely because chemical controls have become ineffective due to resistance to pesticides, and because biological control helps circumvent unwanted side effects of chemical applications, including the destruction of natural enemy communities and environmental pollution (Murphy and La Salle, Reference Murphy and LaSalle1999; Reitz et al., Reference Reitz, Gao, Lei and Tradan2013; Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020). Within Australia, it is important to improve the understanding of the indigenous and adventive parasitoid wasps given the quarantine challenges of deliberately introducing exotic species into the country. In this study, we deployed DNA barcoding and morphological diagnosis to identify 14 parasitoid species which we reared from agromyzids in Australia. Based on our phylogenetic analyses, we found clear genetic divergence within C. pubicornis, D. isaea, H. varicornis, and Opius spp., highlighting the importance of further taxonomic studies on these taxa. In addition, we provide new barcodes with convincing morphological characterization for other species. We also checked for endosymbiont infection status and found both Rickettsia and Wolbachia infections and evidence for a superinfection in one species.
Chrysocharis pubicornis is a koinobiont endoparasitoid of agromyzid leafminers (Lardner, Reference Lardner1991; Baeza Larios, Reference Baeza Larios2007) and is an adventive species in Australia, found in New South Wales, South Australia, Tasmania, and Victoria (Bouček, Reference Bouček1988). It is primarily a pupal parasitoid but occasionally acts as a larval–pupal parasitoid (Hansson, Reference Hansson1985; Lardner, Reference Lardner1991; Baeza Larios, Reference Baeza Larios2007). This species is an important parasitoid wasp of Phytomyza spp. which pupate in the leaf mine (e.g., P. horticola) but may only have a minor impact on Liriomyza pests which generally pupate in the soil (Baeza Larios, Reference Baeza Larios2007; Coquilleau, Reference Coquilleau2020; Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020). In this study, we found 0.2–6.3% COI divergence in C. pubicornis specimens from the same population with haplotypes potentially associated with different host leafmining species. This situation is also found in the UK, where individuals found on P. horticola (BOLD: GBAH8983–14) varied 6.1% from individuals found on Scaptomyza flava (Fallen) (BOLD: GBAH8982–14) (Derocles et al., Reference Derocles, Evans, Nichols, Evans and Lunt2015) (table S3). Additionally, the sympatric genetic divergence of C. pubicornis has been recorded in Norway (where an individual (BOLD: GMNWK1832–14) varied 6.1% from another individual (BOLD: GMNWL1999–14) although the host flies remain unknown. These high and variable levels of sequence divergence in C. pubicornis may indicate cryptic species that require further analysis.
Diglyphus isaea is a synovigenic idiobiont ectoparasitoid of many leafmining Diptera (Zhang et al., Reference Zhang, Liu, Wang, Wan and Li2011) and is released extensively in glasshouses for augmentative biological control of L. sativae, L. trifolii, L. huidobrensis and L. bryoniae (Van Lenteren, Reference Van Lenteren2012). This species has become cosmopolitan after inoculative introductions into other regions (e.g., Japan, Hawaii, Canada, and New Zealand) (Minkenberg, Reference Minkenberg1989; Abe Reference Abe2017). Based on the analyses of COI and ITS1 sequence data, Sha et al. (Reference Sha, Zhu, Murphy, La Salle and Huang2006, Reference Sha, Zhu, Murphy and Huang2007) indicated a probable complex of cryptic species present in Chinese D. isaea.
Derocles et al. (Reference Derocles, Evans, Nichols, Evans and Lunt2015) also suggested D. isaea found in the UK is a species complex due to high intraspecific variability, and our results support this notion. In Australia, D. isaea is an adventive species and possibly introduced from New Zealand, where introductions from Pakistan were released in an attempt to suppress leafminers infesting forage brassicas in the 1970s (McGregor, Reference McGregor, Cameron, Hill, Bain and Thomas1989). In this study, we compared specimens with the samples collected by Sha et al. (Reference Sha, Zhu, Murphy, La Salle and Huang2006, Reference Sha, Zhu, Murphy and Huang2007) and found D. isaea samples from Australia were genetically similar to those from the Netherlands and also clustered with Chinese D. isaea in the largest clade (Group I). Our phylogenetic analyses indicated that Chinese and Australian D. isaea strains are probably the result of the movement of European D. isaea across the world although more populations and individuals are needed to support this hypothesis.
Hemiptarsenus varicornis is a synovigenic idiobiont ectoparasitoid, primarily attacking third instar agromyzid larvae (Bordat et al., Reference Bordat, Coly and Roux-Olivera1995). Host-killing behaviors of this wasp include parasitism, host feeding, and host stinging, which account for 26, 58, and 16% of mortality, respectively (Cheng et al., Reference Cheng, Cao, Zhang, Guo, Wan and Liu2017). Hemiptarsenus varicornis is widely distributed throughout Australia (Bouček, Reference Bouček1988) and its biology is well studied overseas (Bordat et al., Reference Bordat, Coly and Roux-Olivera1995; Thu and Ueno, Reference Thu and Ueno2002; Cheng et al., Reference Cheng, Cao, Zhang, Guo, Wan and Liu2017). In this study, we found all Australian H. varicornis individuals were clustered together, separated from samples from Fiji, Pakistan, Malaysia and Vietnam but nevertheless with relatively low sequence divergence. Prijono et al. (Reference Prijono, Robinson, Rauf, Bjorksten and Hoffmann2004) found H. varicornis in Australia to be more susceptible to abamectin compared with Indonesian H. varicornis. It is possible that insecticide tolerance differs across regions reflecting past histories of chemical selection and/or genetic differences between haplotypes.
Opius spp. are koinobiont larval–pupal endoparasitoids and form one of the largest genera in the family Braconidae (Wharton, Reference Wharton1988). Many Opius species play important roles in the control of leafmining Agromyzidae (Belokobylskij et al., Reference Belokobylskij, Wharton and La Salle2004). For example, in Florida, Opius dissitus Muesebeck (Hymenoptera: Braconidae) was the most abundant parasitoid of L. trifolii on Phaseolus vulgaris L., and a direct density-dependent relationship was detected between O. dissitus parasitism and L. trifolii (Li et al., Reference Li, Seal, Leibee and Liburd2012). A handful of field surveys in Australia demonstrated Opius spp. commonly parasitize agromyzids and might potentially be used to suppress exotic leafmining pests (Lardner, Reference Lardner1991; Bjorksten et al., Reference Bjorksten, Robinson and La Salle2005; Lambkin et al., Reference Lambkin, Fayed, Manchester, La Salle, Scheffer and Yeates2008).
However, knowledge of Opius spp. attacking agromyzids in Australia remains poor since relatively few species are known from rearing records, and many species have no host records (Belokobylskij et al., Reference Belokobylskij, Wharton and La Salle2004). In this study, we found three Opius species with one identified as O. cinerariae and the other two (Opius sp. 1 and Opius sp. 2) identified to genus level. The morphological diagnosis of O. cinerariae is based on the key and re-description in Belokobylskij et al. (Reference Belokobylskij, Wharton and La Salle2004), who examined wasps from L. chenopodii in Australia. In other work, Bjorksten et al. (Reference Bjorksten, Robinson and La Salle2005) found O. cinerariae from L. chenopodii on Beta vulgaris and Lardner (Reference Lardner1991) found O. cinerariae attacking L. brassicae on Brassica napus, Raphanus raphanistrum, Raphanus rugosum, and Sisymbrium officinale. Belokobylskij et al. (Reference Belokobylskij, Wharton and La Salle2004) examined many Australian specimens, including the holotype from Queensland, and noted that O. cinerariae is more variable than the original description by Fischer (Reference Fischer1963). In this study, the genetic divergence of O. cinerariae specimens between Australia and New Zealand ranged from 6.9–8.0%. Further collections of this species in other regions are needed to determine if there is a cryptic species complex. Opius sp. 1 and Opius sp. 2 are more genetically similar than O. cinerariae but the divergence between the two is still substantial. Apart from O. cinerariae, Lardner (Reference Lardner1991) collected Opius atricornis from L. brassicae in South Australia. Further studies are needed to check if our specimens of Opius sp. 1 and Opius sp. 2 are in fact Opius atricornis (Belokobylskij et al., Reference Belokobylskij, Wharton and La Salle2004).
The Victorian Aprostocetus (Aprostocetus) sp. belongs to the subfamily of Tetrastichinae (Graham, Reference Graham1987; LaSalle, Reference LaSalle1994). In Australia, 207 Aprostocetus species have been described but no doubt many more species remain undescribed (Bouček, Reference Bouček1988). Three Aprostocetus species (spp. 1, 2 and 3) were reared from P. praecellens on R. candolleana and one species (sp. 2) from R. parabolica in South Australia (Wood et al., Reference Wood, Siekmann, Stephens, DeGraaf, La Salle and Glatz2010), but it is not known whether the Victorian Aprostocetus (Aprostocetus) sp. reared from P. praecellens on R. parabolica is the same as Aprostocetus (sp. 2). Further molecular and morphological work will be essential to unravel the identity of the Aprostocetus species attacking Australian agromyzids given this is a very large and taxonomically diverse genus, with a large number of undescribed species.
Asecodes is a small genus with a cosmopolitan distribution (Noyes, Reference Noyes2019). To date, all Asecodes species reared from agromyzids were originally considered to be Teleopterus. Bouček (Reference Bouček1988) noted that there were 3–4 species of Teleopterus in Australia, with only T. atripes (Girault, Reference Girault1915a) described from Queensland, but without host data. Subsequently, Teleopterus was synonymized with Asecodes (Hansson, Reference Hansson1996). Gumovsky (Reference Gumovsky2001) then synonymized Asecodes, Neochrysocharis, Hispinocharis, and Mangocharis with Closterocerus. However, molecular analysis led Burks et al. (Reference Burks, Heraty, Gebiola and Hansson2011) to remove Neochrysocharis and Asecodes from synonymy. Asecodes delucchii and A. erxias are two common parasitoids of agromyzid wasps and may play an important role in suppressing Liriomyza pests and P. horticola (Arakaki and Kinjo, Reference Arakaki and Kinjo1998; Tran et al., Reference Tran, Tran, Konishi and Takagi2005; Amano et al., Reference Amano, Suzuki, Hiromori and Saito2008; Tran Reference Tran2009). In this study, we only identify Asecodes to genus level and DNA sequences demonstrate our Asecodes sp. is not A. erxias (Genbank: MG836471.1/MG836472.1) or other Asecodes species present in BOLD and GenBank. Coquilleau (Reference Coquilleau2020) showed the Asecodes sp. sequenced in this study is commonly found parasitizing L. brassicae, P. plantaginis, and P. syngenesiae in Melbourne and we have also found this Asecodes sp. regularly reared from L. huidobrensis in Queensland (P. Ridland, unpub. data).
Closterocerus mirabilis is an idiobiont ectoparasitoid but little is known about its biology (Lardner, Reference Lardner1991; Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020). It has been found in ACT, South Australia, Victoria and Queensland (Edwards and La Salle, Reference Edwards and La Salle2004; Bjorksten et al., Reference Bjorksten, Robinson and La Salle2005; Lambkin et al., Reference Lambkin, Fayed, Manchester, La Salle, Scheffer and Yeates2008; Coquilleau, Reference Coquilleau2020) and is suspected to be present in Indonesia (Edwards and La Salle, Reference Edwards and La Salle2004). Closterocerus mirabilis is morphologically close to Closterocerus cruy (Girault), but the former has only been recorded from agromyzid leafminers whereas the latter has only been recorded from lepidopteran hosts (Berry, Reference Berry2007a). Both these species are morphologically very similar to Closterocerus separatus Li & Li (Li and Li Reference Li and Li2021). Closterocerus mirabilis has been reared from P. plantaginis, P. syngenesiae, L. chenopodii, L. brassicae, and L. sativae (Edwards and La Salle, Reference Edwards and La Salle2004; Bjorksten et al., Reference Bjorksten, Robinson and La Salle2005; Lambkin et al., Reference Lambkin, Fayed, Manchester, La Salle, Scheffer and Yeates2008; Coquilleau, Reference Coquilleau2020; Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020) and is an abundant indigenous parasitoid of agromyzid leafminers in eastern Australia. We found only a single haplotype in this study.
Dacnusa areolaris is a koinobiont endoparasitoid, which oviposits in the early larval stages and emerges from the puparia of agromyzid hosts (Haviland, Reference Haviland1922). This species is thought to have been accidentally introduced into Australia and New Zealand (Wharton and Austin, Reference Wharton and Austin1991; Berry, Reference Berry2007b) and no native Dacnusa species have been found in Australia to date (Wharton and Austin, Reference Wharton and Austin1991). In Australia, Dac. areolaris have been recorded from South Australia, Victoria, Tasmania, the Australian Capital Territory, and New South Wales; the earliest collection records are from 1927 (Wharton and Austin, Reference Wharton and Austin1991). Griffiths (Reference Griffiths1966) recorded Dac. areolaris reared from three hosts including P. syngenesiae, Phytomyza asteris (Hendel), and Phytomyza nigra (Meigen). Similar to Cl. mirabilis, we only found a single haplotype of Dac. areolaris in this study.
Neochrysocharis formosa and N. okazakii are synovigenic idiobiont endoparasitoids (Chien and Chang, Reference Chien and Chang2009a, Reference Chien and Chang2009b). Both species are widely distributed in Asian countries and parasitize Liriomyza (Tran et al., Reference Tran, Ueno and Takagi2007; Sunari et al., Reference Sunari, Supartha, Wijaya and Laba2016). In Australia, Bjorksten et al. (Reference Bjorksten, Robinson and La Salle2005) recorded one specimen of N. okazakii on L. brassicae and one specimen of an unidentified Neochrysocharis sp. collected from P. syngenesiae. Lambkin et al. (Reference Lambkin, Fayed, Manchester, La Salle, Scheffer and Yeates2008) found unidentified Neochrysocharis sp. from P. plantaginis and P. syngenesiae, and field collections in Victorian have found N. formosa reared from L. brassicae, L. chenopodii, P. plantaginis and P. syngenesiae (Coquilleau, Reference Coquilleau2020; Xu et al., Reference Xu, Hoffmann, Umina, Coquilleau, Gill and Ridland2022). Currently, only female wasps have been found in Australia and thelytokous strains of N. formosa have been recorded in Japan and China which are associated with Rickettsia infection (Hagimori et al., Reference Hagimori, Abe, Date and Miura2006; Zhang et al., Reference Zhang, Lu, Liu, Wang, Wang and Wan2014; Yang et al., Reference Yang, Xuan, Ye, Guo, Yang and Liu2017; Xu et al., Reference Xu, Hoffmann, Umina, Coquilleau, Gill and Ridland2022). For N. okazakii, only limited DNA barcodes are publicly available. Our 28S sequence of N. okazakii is identical to N. okazakii (NCBI: AB526861) from Japan (Adachi-Hagimori et al., Reference Adachi-Hagimori, Miura and Abe2011), but there is still 6.3% genetic distance from Chinese N. okazakii based on 430 bp COI sequence data (Personal Communication W-X Liu). We suspect there are cryptic species within this taxon despite not finding any morphological differences through SEM imaging.
Trigonogastrella parasitica is a larval–pupal koinobiont endoparasitoid of agromyzids and has been found in Queensland, New South Wales, ACT, Victoria, and Tasmania (Bouček, Reference Bouček1988). The species was described by Girault (Reference Girault1915b) from Victorian specimens together with another species, Cryptoprymnoides rabiosus from Queensland, which was subsequently re-classified as Trigonogastrella rabiosa (Girault) (Bouček, Reference Bouček1988). Trigonogastrella rabiosa has been recorded in Queensland, New South Wales, South Australia, and Western Australia without detailed host information (Bouček, Reference Bouček1988). The known agromyzid hosts of T. parasitica include P. syngenesiae, Liriomyza spp., Ophiomyia spp., and P. plantaginis (Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020). Limited field surveys illustrated that T. parasitica is a common parasitoid species reared from P. syngenesiae and P. plantaginis (Lambkin et al., Reference Lambkin, Fayed, Manchester, La Salle, Scheffer and Yeates2008; Coquilleau, Reference Coquilleau2020) and we found at least three COI haplotypes exist in Australia.
Zagrammosoma latilineatum was first described in Indonesia and Australia and has been found parasitizing L. huidobrensis in southeast Asia (Ubaidillah et al., Reference Ubaidillah, LaSalle and Rauf2000). This idiobiont ectoparasitoid species is widely distributed in Australia, recorded in every state except for Tasmania (Lardner, Reference Lardner1991; Perry and Heraty, Reference Perry and Heraty2021). Bjorksten et al. (Reference Bjorksten, Robinson and La Salle2005) reared Z. latilineatum from L. brassicae and L. chenopodii in Victoria. Wood et al. (Reference Wood, Siekmann, Stephens, DeGraaf, La Salle and Glatz2010) found that Z. latilineatum was commonly reared from Phytoliriomyza praecellens in South Australia. It was also the most frequently reared species from L. sativae on Thursday Island, Horn Island and Seisia between 2018 and 2019 (Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020). Given Z. latilineatum is widely distributed and can inflict high rates of parasitism on exotic Liriomyza pests (Ridland et al., Reference Ridland, Umina, Pirtle and Hoffmann2020), this species is considered a potentially important biological control agent for the management of Liriomyza pests in Australia.
Endosymbiont assays in the present study found H. varicornis was infected with Wolbachia at a low frequency in Victoria, whereas all individuals from Fiji were infected with Wolbachia. Both H. varicornis and its host L. brassicae at the Victorian collection site (Flemington Bridge) were infected by Wolbachia with identical wsp sequences. Likewise, Tagami et al. (Reference Tagami, Doi, Sugiyama, Tatara and Saito2006) found nearly identical wsp sequences in H. varicornis and L. bryoniae. In Japan, Wolbachia infection did not induce cytoplasmic incompatibility (CI) or parthenogenesis in H. varicornis. We also found Opius sp. 1 and Opius sp. 2 infected with Wolbachia at low frequencies. The wsp sequences of Opius sp. 1 and D. isaea are identical to the wsp sequences of L. sativae (wLsatC: MW310402), P. syngenesiae (MW047083) and P. praecellens (MW310408) (Xu et al., Reference Xu, Ridland, Umina, Gill, Ross, Pirtle and Hoffmann2021a). This suggests horizontal transmission of Wolbachia may occur through host-parasitoid interactions. Moreover, we found all Cl. mirabilis individuals in this study are infected with Wolbachia but it is unknown if there are any phenotypes associated with this Wolbachia strain.
We found three wasp species infected with Rickettsia including one species with Wolbachia/Rickettsia co-infections. In N. formosa, only one COI haplotype (NF_5COI.03) was found in Queensland and this is a single base pair different to N. formosa collected in Victoria (NCBI: OK076721, NF_5COI.02). Interestingly, NF_5COI.02 and another N. formosa haplotype collected at the same location, NF_5COI.01 (NCBI: OK076720), were found to be thelytokous (Xu et al., Reference Xu, Hoffmann, Umina, Coquilleau, Gill and Ridland2022). Given no males of N. formosa were collected from Queensland in this study and all specimens were infected with Rickettsia, it is possible that NF_5COI.03 is also thelytokous. In N. okazakii, we found all individuals (three females and three males) are not only infected with Rickettsia (same as found in N. formosa, NCBI: OK086364.1) but also infected with Wolbachia (same as found in L. brassicae, NCBI: MW047082.1). The Rickettsia found in N. okazakii was not associated with thelytoky but it has been suggested to cause thelytoky in N. formosa (Hagimori et al., Reference Hagimori, Abe, Date and Miura2006). The Wolbachia strain in N. okazakii has been suggested to cause cytoplasmic incompatibility in L. brassicae (Xu et al., Reference Xu, Ridland, Umina, Gill, Ross, Pirtle and Hoffmann2021a). It would be worthwhile exploring how these two endosymbionts interact in N. okazakii and if there are any phenotypic effects detected. For Aprostocetus sp., only females were collected in this study and all individuals tested were infected with Rickettsia with two base pairs different from Rickettsia detected in N. formosa and N. okazakii.
Although the total number of wasps tested was somewhat limited, endosymbiont infection status appears to be associated with the mtDNA haplotypes. Species with diverse COI haplotypes were often infected with a low frequency of Wolbachia or were devoid of Wolbachia, whereas species infected with Wolbachia at a high frequency (e.g., Cl. mirabilis in Australia, H. varicornis in Fiji) often possess fewer haplotypes. A decrease of mitochondrial variation is thought to relate to Wolbachia-induced cytoplasmic incompatibility, which results in the infected individuals spreading in a population and eventually throughout the whole range of a species (Hale and Hoffmann, Reference Hale and Hoffmann1990).
Conclusion
Overall, our study provides important DNA barcodes and morphological information of 14 parasitoid wasp species that are potentially important in agromyzid control in Australia. Given the taxonomic challenges and limited DNA barcoding information available on public databases, our study provides a solid foundation that facilitates future research into agromyzid wasps. Furthermore, our study provides information on endosymbiont infections across parasitoid species, with the potential of manipulating endosymbionts to alter the mode of reproduction in populations of the parasitoids in the future.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485323000160
Acknowledgments
We thank Christer Hansson for his advice on Asecodes sp. and Neochrysocharis okazakii; Kees van Achterberg for his advice on Opius taxonomy; Wanxue Liu for his advice on Chrysocharis pubicornis and Neochrysocharis okazakii; John Duff for parasitoid samples from Queensland; Markus Knapp for the sample of Diglyphus isaea from the Koppert BV culture; Zhixing Lin and Roger Curtain for technical advice and assistance with SEM at the Bio21 Advanced Microscopy Facility (The University of Melbourne). We acknowledge Qiong Yang and Nancy Endersby-Harshman for technical advice and assistance with molecular experiments. We appreciate the thoughtful comments provided by two anonymous reviewers. This project was supported by the RD&E program for control, eradication and preparedness for vegetable leafminer (MT16004) and the management strategy for serpentine leafminer (Liriomyza huidobrensis) (MT20005), funded by Hort Innovation. Xuefen Xu acknowledges support from the David Hay Fund.
Conflict of interest
The author(s) declare no conflicts of interest.











