A genome-wide association study (GWAS) of schizophrenia (SZ) risk, which included almost 48,000 individuals (12,495 cases and 34,951 controls), identified the single nucleotide polymorphism (SNP) rs12807809 (C/T) as showing genome-wide significance, p = 2.14 × 10−9; OR = 1.15, 95% CI [1.1, 1.2] (Stefansson et al., Reference Stefansson, Ophoff, Steinberg, Andreassen, Cichon, Rujescu and Collier2009). This SNP is located 3,457 bases upstream of the neurogranin (NRGN) gene, which has an integral role in the calcium-calmodulin signaling pathway (Hayashi, Reference Hayashi2009). The NRGN protein is expressed exclusively in the brain in humans (Martinez de Arrieta et al., Reference Martinez de Arrieta, Perez Jurado, Bernal and Coloma1997) and is the primary postsynaptic protein in the regulation of calmodulin availability in neurons, since it will bind to calmodulin in the absence of calcium (Baudier et al., Reference Baudier, Deloulme, Van Dorsselaer, Black and Matthes1991). With regard to regional specificity, NRGN is expressed profusely in regions of the brain that are important for learning and memory, such as the CA1 pyramidal cells of the hippocampus (Huang et al., Reference Huang, Huang and Boucheron2007), and NRGN knockout mice show notable deficits in behavioral measures that are reliant on hippocampal function (Huang et al., Reference Huang, Huang, Jager, Li, Reymann and Balschun2004). Furthermore, reductions in NRGN immunoreactivity in prefrontal regions have been observed in postmortem samples from SZ patients (Broadbelt & Jones, Reference Broadbelt and Jones2008; Broadbelt et al., Reference Broadbelt, Ramprasaud and Jones2006). Collectively, these data suggest a role for NRGN/rs12807809 in functional and structural variability in the hippocampus. Given the role of this region in cognitive function, it is probable that the impact of rs12807809 on hippocampal function and structure contributes to altered cognitive function in SZ.
To date, there have been three investigations of the effects of this NRGN risk variant on neurocognitive intermediate phenotypes for schizophrenia. Our own study of the impact of NRGN genotype on cognitive and/or behavioral function in samples of 393 cases and 157 controls failed to find any evidence of a significant association between rs12807809 and performance on a range of neuropsychological measures (Donohoe et al., Reference Donohoe, Walters, Morris, Da Costa, Rose, Hargreaves, Maher, Hayes, Giegling, Hartmann, Möller, Muglia, Moskvina, Owen, O'Donovan, Gill, Corvin and Rujescu2011). Conversely, investigations of the functional consequences of this variant have been more fruitful. For example, Krug et al., (Reference Krug, Krach, Jansen, Nieratschker, Witt, Shah, Nöthen, Rietschel and Kircher2011) found that healthy individuals who were homozygous carriers of the risk T allele exhibited differential function in the anterior and posterior cingulate during performance of an episodic memory task, compared to those with one or two copies of the non-risk C allele. Similarly, Pohlack et al. (Reference Pohlack, Nees, Ruttorf, Witt, Nieratschker, Rietschel and Flor2011) observed a relative reduction in hippocampal function in homozygous T individuals, compared to C carriers during the acquisition phase of a contextual fear paradigm.
Collectively, these studies suggest that, although the impact of NRGN/rs12807809 may not be apparent in cognitive or behavioral performance, indices of brain integrity may prove useful in the delineation of the contribution of this variant to SZ risk. Therefore, the aim of the current investigation was to ascertain whether the functional changes attributable to NRGN genotype in healthy volunteers in measures of episodic memory function extended to other cognitive processes (i.e., working memory/executive function). Furthermore, we sought to determine whether the impact of rs12807809 was apparent in variability in brain structure.
Methods
Participants
Structural Imaging (sMRI). Individuals were selected from the Trinity College Institute for Neuroscience imaging biobank project, which involved the opportunistic sampling of all healthy, right-handed controls participating in magnetic resonance imaging (MRI) studies at the Institute. Participants gave consent to the use of structural data collected under the primary study and provided a saliva sample for genetics analysis. From this sample, 140 individuals (63 male; mean age = 27.60 years; mean years education = 16.93; see Table 1 for full details) were included, based upon the quality of T1 structural data and successful genotyping of the NRGN (rs12807809) variant. Forty-eight structural imaging (sMRI) participants also participated in functional imaging (fMRI).
TABLE 1 Summary of Participant Demographic Data
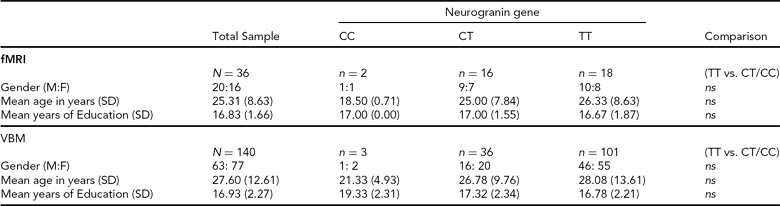
SD = standard deviation, M:F = males:females, ns = not statistically significant, fMRI = functional magnetic resonance imaging, VBM = voxel-based morphometry.
The majority of participants (N = 97) were confirmed as Irish (i.e., having Irish-born maternal and paternal grandparents). Lineage information was not available for the remaining participants (N = 43). However, given the relative homogeneity of the Irish population (e.g., 92% of Dublin city population is Irish/Caucasian; www.cso.ie/census), the vast majority of these participants were also likely to have been of Irish lineage. Furthermore, this subgroup did not differ from known Irish participants in terms of genotype frequency, gender, age, or voxel-based morphometry (VBM) analysis of grey matter (GM) and white matter (WM) volume.
Functional Imaging (fMRI). Healthy, right-handed Irish individuals (N = 52) were recruited from the general population through local media advertising. In contrast to sMRI participant selection, recruitment advertising for fMRI specified that participants should have both paternal and maternal grandparents who were born in Ireland. This information was confirmed with individuals prior to consent. For the purposes of imaging analysis, we were able to identify 18 individuals who were carriers of the non-risk C allele for NRGN (10 male; mean age = 24.27 years; mean years education = 17.00; see Table 1 for full details). A further 18 individuals (10 male; mean age = 26.33 years; mean years education = 16.67) who were homozygous for the risk T allele and who provided age, gender, and education matches for the C carriers were also included in functional imaging analysis. Participants provided written, informed consent, in accordance with local ethics committee guidelines. Exclusion criteria included relevant neurological, medical or psychiatric history, family history of psychosis (i.e., one or more first-degree relative with a confirmed diagnosis of SZ or other psychosis), claustrophobia, pregnancy, and any other contraindication for MRI.
Procedure
Genotyping. Genetic analysis was carried out using DNA obtained from saliva samples that were collected using Oragene DNA self-collection kits (DNA Genotek). The rs12807809 SNP was genotyped using a Taqman® SNP Genotyping Assay on a 7900HT Sequence Detection System (Applied Biosystems). The call rate for the Taqman genotyping was >95% and the samples were in Hardy-Weinberg Equilibrium (p > 0.05). In addition, a small number of HapMap CEU DNA samples (www.hapmap.org) were genotyped for rs12807809 for quality control purposes and were all found to be concordant with available online HapMap data for this SNP. Genotype frequencies for fMRI and sMRI are noted in Table 1.
MRI. Participants were imaged on a Philips Intera Achieva 3T MR system. Whole-brain BOLD EPI consisting of 32 non-contiguous, axial 3.5 mm slices was acquired with a 0.35 mm slice gap and the following imaging parameters: TR = 2000 ms; TE = 35 ms; FOV = 224×224 mm at 64 × 64 matrix; and flip angle = 90°. The duration of functional scanning was 220TRs/440s. Structural imaging involved the acquisition of a T1-weighted image (180 slices; duration = 6 min) using a TFE gradient echo pulse sequence, with a slice thickness of 0.9 mm and a 230 × 230 mm FOV.
Spatial Working Memory Paradigm. In this block-design task, participants determined whether the spatial location, relative to a white fixation cross, of a white dot (i.e., target) and a red circle (i.e., probe) were either the same (match) or different (no match). The target and probe were each presented for 500 ms. There were three levels/conditions in the task: no delay (baseline); 1-dot; and 3-dot. In the no delay condition, the target was a single white dot, and both the target and probe appeared simultaneously (Figure 1A). During the 1-dot condition (Figure 1B), the target and probe were separated by a 3-second delay. Similarly, the 3-dot condition (Figure 1C) also incorporated a 3-second inter-stimulus interval; however, in this condition, the target image was comprised of three dots. During the 3-dot condition, the probe was also a single red circle and participants were asked to judge whether the probe was in the same position as any one of the three dots in the image that preceded it.
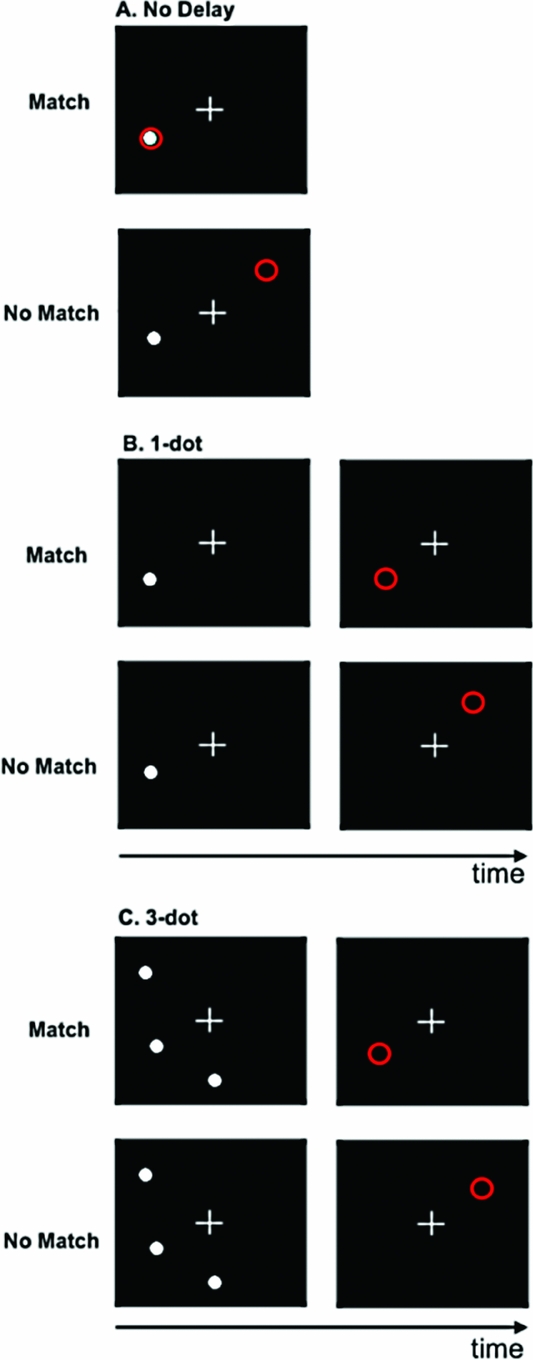
FIGURE 1 Spatial working memory task. A. No delay; B. 1-dot; C. 3-dot.
There were six trials per block with an inter-trial interval of 2 s. Participants completed four blocks of each condition (i.e., 12 blocks/72 trials total). The order of blocks was fixed; however, to account for the potentially confounding effect of block order on brain activity across the session, there were two predetermined block orders and participants were pseudo-randomly allocated to one or the other prior to imaging. Participants were given left-hand and right-hand MRI-compatible response units. They responded with a left button press for a match and a right button press for no match. Behavioral measures of interest on the spatial working memory (SWM) task were accuracy (i.e., number of correctly identified trials) and reaction time (RT).
Data Analysis
fMRI. Functional imaging data were analyzed using Analysis of Functional Images (AFNI; Cox, Reference Cox1996). To correct for head motion, 3D EPI data for each subject were co-registered to a base volume. The subsequent data were inspected for motion using the censor.py application for AFNI (http://brainimaging.waisman.wisc.edu/~perlman/code/censor.py). Using strict censoring criteria (i.e., translation >0.3 mm or rotation >0.3° between consecutive TRs), TRs exceeding these parameters were recorded and were removed from further analysis prior to deconvolution. Data quality control also included the application of an edge detection algorithm to exclude activations occurring outside the brain.
Voxel-wise multiple regression was conducted, in which imaging regressors were expressed as a delta function relative to baseline and convolved with a hemodynamic response function. There were regressors representing the SWM conditions (1-dot and 3-dot) and six motion parameters that were included as regressors of no interest. A voxel-wise average amplitude change (β) equal to the percentage change from baseline was calculated for each SWM condition. The resultant activation maps for each subject were then registered to a higher resolution (1 μl) standard space (Talairach & Tournoux, Reference Talairach and Tournoux1988) and spatially blurred using a 4.1 mm Gaussian isotropic kernel.
Whole-brain mixed measures ANOVA were used to consider the impact of genotype group and SWM condition (1-dot vs. 3-dot) on brain activity. In light of the relatively small number of C homozygotes, analyses considered relative differences in homozygous risk carriers (i.e., TT) vs. carriers of the non-risk C allele (i.e., CT & CC). These groups were matched for age, gender, and years of education (Table 1). A voxel-wise threshold correcting for multiple comparisons and controlling for family-wise error (FWE) rate was calculated using a Monte Carlo simulation. This deterministic sampling algorithm ascertains the frequency of significant clusters that would occur by chance under the null hypothesis (i.e., the false positive rate). Using an iterative process that involves random image generation, Gaussian filtering, thresholding, image masking, and cluster size frequency, the AlphaSim program in AFNI was used to generate an estimate of the overall significance level for various combinations of cluster extent and probability thresholds (for full details on this program see afni.nimh.nih.gov/afni/doc/manual/AlphaSim). Cluster significance was determined as equaling or exceeding a given cluster extent threshold (i.e., main effect of genotype = 414 voxels; main effect of SWM load = 889 voxels; gene x SWM = 235 voxels) and height threshold of p < .001 with p < .05 cluster threshold for FWE following correction for multiple comparisons at the whole brain level. The direction of difference in those clusters showing a main effect of genotype or working memory load was confirmed using a priori contrasts, which were also corrected for whole-brain multiple comparisons.
Voxel-Based Morphometry. For an explanation of voxel-based morphometry, see Ashburner and Friston (Reference Ashburner and Friston2000). sMRI analysis was performed within SPM5 (http://www.fil.ion.ucl.ac.uk/spm) running under Matlab (v7.8; The MathWorks) and utilizing the VBM toolbox (v5.1; http://dbm.neuro.uni-hen.de/vbm). Individual volumes were visually inspected for scanner artifacts and gross anatomical abnormalities. Volumes that passed initial data quality control were segmented into GM, WM and cerebrospinal fluid, without tissue priors and using a Hidden Markov Random Field weighting of 0.15. Segmented images were normalized using the DARTEL toolbox (Ashburner, Reference Ashburner2007), in which GM and WM templates were created using standard parameters. Jacobian scaled (‘modulated’) warped tissue classes were subsequently created for both GM and WM for each subject. The resultant images were smoothed with an 8 mm3 Gaussian kernel.
As with fMRI analyses, sMRI analyses involved mixed model ANOVA, which considered the impact of NOS1 genotype on GM and WM volume. Our analyses focused on the data in native space (i.e., normalized to the DARTEL template) rather than standard space (e.g., MNI template). Given the variability in age (range = 18–75 years), our sample included individuals from a range of developmental stages, and we felt that normalization to a standard adult template would not be appropriate. Modeling included genotype as a fixed factor with two levels (i.e., CC & CT vs. TT). Furthermore, although the groups were essentially matched for age, gender, and years of education (Table 1), in order to account for normal variation in brain structure, we included age, gender, and total GM or WM volume as covariates. Significance was determined as the voxel-wise threshold at p CORRECTED(FWE) < .05, following correction for multiple comparisons at the whole-brain level. Because nonuniform smoothness of VBM data can influence interpretation of these types of analyses (Ashburner & Friston, 2000; Worsley et al., Reference Worsley, Andermann, Koulis, MacDonald and Evans1999), determination of significance included a non-stationarity cluster extent correction, which utilized the random field theory version of cluster inference under non-stationarity (Hayasaka et al., Reference Hayasaka, Phan, Liberzon, Worsley and Nichols2004) and was implemented using the NS toolbox (http://fmri.wfubmc.edu/cms/NS-General),
Other Data. Behavioral and demographic data were analyzed in SPSS (v16; SPSS Inc. Chicago, IL) and focused on the relative differences between genotype groups.
Results
SWM Behavioral Data
With regard to behavioral performance, there was a significant main effect of working memory load on both accuracy, F(2,66) = 18.30, p < .001, and RT, F(2,66) = 8.56, p < .001, but no effect of genotype group nor genotype by load interactions. Post hoc calculations of effect size indicated that genotype accounted for less than 2% of total variability in behavioral performance (i.e., accuracy: partial η2 = 0.016; RT: partial η2 = 0.019).
sMRI
There was no significant effect of NRGN genotype on either GM or WM volume following correction for multiple comparisons (i.e., voxelwise p (FWE) < .05). Even at the more lenient uncorrected threshold of p < .001 and a minimum cluster extent of 100 voxels, we failed to observe any significant effect of genotype on brain structure.
fMRI
Main Effect of Working Memory Load on Brain Activity. Load-dependent changes in activity were noted in the right middle and left inferior occipital gyri, the left superior parietal cortex, cuneus, and posterior cingulate, and bilaterally in BA6 (Table 2). In the left cuneus and posterior cingulate, there was a load-dependent decrease in activity (i.e., 1-dot > 3-dot). All other clusters showing a main effect of load exhibited load-dependent increases (i.e., 1-dot < 3-dot; Table 2).
TABLE 2 Main Effect of Spatial Working Memory Maintenance Condition on Brain Activity
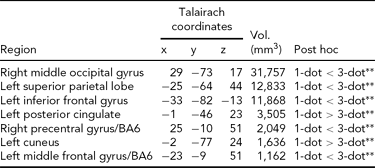
All regions shown are significant at p CORRECTED < .05 (minimum cluster extent = 890 voxels); 1-dot and 3-dot = spatial working memory conditions.
** =p < .001.
Effect of Genotype. There was a main effect of genotype (i.e., CC/CT < TT) in a single cluster in the left superior frontal gyrus/BA10 (Talairach co-ordinates: -14 61 8; cluster extent = 474 voxels/mm3; p CORRECTED < .05; Figure 2). There were no other regions showing a main effect of genotype, and no genotype x load interaction effects on brain activity.

FIGURE 2 Main effect of neurogranin (rs12807809) genotype on brain activity in ventromedial prefrontal cortex during performance of the spatial working memory paradigm. Note: F ≥ 13.03, p CORRECTED < .05; minimum cluster extent = 414 voxels/mm3; cluster rendered on the ICBM452 T1 template from AFNI (Analysis of Functional NeuroImages); graph shows mean signal change within each genotype group (CC/CT vs. TT) averaged across working memory conditions; **contrast p < .001.
Discussion
The genome-wide associated SZ risk variant rs12807809 at NRGN, previously shown to be associated with variability in brain activity associated with episodic memory (Krug et al., Reference Krug, Krach, Jansen, Nieratschker, Witt, Shah, Nöthen, Rietschel and Kircher2011; Pohlack, et al., 2011), was shown here to be related to functional changes in the ventromedial prefrontal cortex (vmPFC) in healthy controls during performance of an SWM task. Specifically, while carriers of one or two copies of the non-risk C allele seemed to disengage this region during performance of the task, homozygous T risk carriers failed to significantly reduce activation, irrespective of memory load. Conversely, NRGN genotype was not associated with variability in behavioral performance or brain structure.
It is intriguing that the main effect of NRGN genotype was seen in the vmPFC. The medial prefrontal cortex (mPFC) has been implicated in SZ-mediated changes in executive function. For example, Pomarol-Clotet et al. (Reference Pomarol-Clotet, Salvador, Sarro, Gomar, Vila, Martinez, Guerrero, Ortiz-Gil, Sans-Sansa, Capdevila, Cebamanos and McKenna2008) found that, during performance of an n-back task, patients with SZ showed reduced deactivation in medial frontal regions. Moreover, a recent structural study of the correlates of dysexecutive function in SZ found that reduced GM volume in mPFC was associated with executive dysfunction in patients (Kawada et al., Reference Kawada, Yoshizumi, Hirao, Fujiwara, Miyata, Shimizu, Namiki, Sawamoto, Fukuyama, Hayashi and Murai2009). Other studies considering alternative measures of SWM (e.g., mental rotation) have suggested that differences between cohorts in task-related changes in BOLD signal in the absence of differences in performance are indicative of alterations in processing efficiency (e.g., Christova et al., Reference Christova, Lewis, Tagaris, Ugurbil and Georgopoulos2008). Therefore, that we observed risk-mediated increases in mPFC activity in the absence of significant behavioral differences between groups suggests that the mechanism by which NRGN/rs12807809 confers risk for SZ may, in part, be attributable to a genotype-mediated reduction in processing efficiency in mPFC, or networks that involve this region.
The mPFC is also implicated in the default mode network (DMN), that is, those regions involved in self-referential mental activity and which normally show reduced activity during goal-directed behaviors (Buckner & Vincent, Reference Buckner and Vincent2007; Buckner et al., Reference Buckner, Andrews-Hanna and Schacter2008; Gusnard & Raichle, Reference Gusnard, Akbudak, Shulman and Raichle2001, Gusnard et al., Reference Gusnard and Raichle2001; Raichle et al., Reference Raichle, MacLeod, Snyder, Powers, Gusnard and Shulman2001). A growing body of literature suggests abnormal DMN function in patients with SZ (see Broyd et al., Reference Broyd, Demanuele, Debener, Helps, James and Sonuga-Barke2009 for review), with patients showing a range of DMN abnormalities. Furthermore, it has been postulated that variability in SZ susceptibility genes contributes to dysfunctional activity during cognitive processing via an impact on the coordination of functional networks in patients (Tan et al., Reference Tan, Callicott and Weinberger2007). The failure of homozygous T carriers to disengage vmPFC to the same extent as C carriers during goal-directed behavior may, therefore, be indicative of a role for NRGN/rs12807809 in DMN abnormalities in SZ. However, it is important to note that, due to the absence of a true rest condition, our paradigm does not allow us to determine whether carriers of the risk T allele show changes in DMN function or connectivity. Delineating whether alterations in DMN function constitute a significant role of the rs12807809 variant in SZ risk will require explicitly testing whether differences are notable between risk and non-risk individuals at rest.
The lack of a significant effect of genotype on working memory performance (accuracy and/or RT) is consistent with our previous work and that of others, which failed to demonstrate NRGN-specific effects on cognitive performance (Donohoe, et al., 2011; Krug, et al., 2011). While in the current investigation this might be attributed to significantly reduced power to detect cognitive effects, effect size estimates suggest that genotype accounted for less than 2% of variance in performance measures. Therefore, even with much greater numbers, we do not anticipate that T homozygous individuals would exhibit performance deficits on this particular SWM measure.
In line with previous observations suggesting a lack of influence of rs12807809 on hippocampal volume (Pohlack et al., Reference Pohlack, Nees, Ruttorf, Witt, Nieratschker, Rietschel and Flor2011), our data were also indicative of no impact of this variant on GM or WM volume. While it could be argued that the absence of structural effects were due to sample-size-related power issues, the lack of any significant effects of genotype in either direction (i.e., CC/CT > TT or CC/CT < TT), even at very lenient significance criteria, suggests that this was not the case. An alternative explanation is that the methodology used to consider brain structure was not sufficiently sensitive to variability in rs12807809 genotype. Although we would anticipate that measures of GM volume, such as VBM, would be sensitive to genetic factors, there is evidence to suggest that alternative methods (e.g., cortical thickness, surface area) may be preferable in determining genotype-related variability in brain structure (Winkler et al., Reference Winkler, Kochunov, Blangero, Almasy, Zilles, Fox, Duggirala and Glahn2010). This potentially limiting factor should be taken into consideration in the interpretation of our results, and should be considered in future sMRI studies of both the rs12807809 SNP and other psychosis risk variants.
The observations presented here are limited by a number of factors. Firstly, the relatively small number of C homozygotes (i.e., sMRI n = 3; fMRI n = 2) required grouping all C carriers into one analysis cohort; thus we were unable to determine any dose-related effects of the rs12807809 risk T allele. Although the relative numbers of C-carrying individuals was representative of other investigations of this NRGN SNP, it is imperative that future studies attempt to better delineate whether there are any likely dose effects of the T allele.
The power of our fMRI analysis is also a concern in the interpretation of our observations. Again, due to the relatively low frequency of the C allele, we were only able to identify 18 individuals who carried one or two copies of the non-risk allele. In an attempt to minimize the impact of other non-genotypic factors that might contribute to brain activity associated with executive function, we were careful to select individuals who provided very close age, gender, and education matches for our participants. Although we were able to select 18 T homozygotes who provided an excellent match for the C cohort, this meant that our total functional imaging analysis group was only 36 individuals — a relatively small number for an imaging genetics investigation. Although we are assured of the reliability of our results due to the rigorous selection of subjects and treatment of the data, replication in a large group is essential in order to confirm the impact of NRGN in ventromedial prefrontal function.
Finally, as with other imaging investigations of the NRGN rs12807809 SNP, our study included only healthy individuals. Although this has the advantage of exploring the influence of NRGN on brain structure and function in the absence of patient-specific cofounding variables (e.g., medication, epistasis with other risk variants, etc.), future exploration of the influence of this SNP on variability in brain function in SZ patients will prove critical in delineating the role of NRGN in the psychosis disease phenotype.
In sum, the data presented here suggest that, while the SZ risk variant rs12807809 is not associated with variability in behavior or brain structure in healthy individuals, it may contribute to altered processing efficiency during executive function via an impact on activity in the vmPFC. In light of the role of vmPFC in the DMN, and DMN abnormalities in SZ reported in the extant literature, this suggests that altered activity in this region in homozygous risk carriers may provide a functional basis through which NRGN may contribute to the SZ disease phenotype.
Acknowledgments
We would like to thank all those who participated in these studies and the staff and students of TCIN who contributed to the biobank project, including Sojo Joseph, Simon Dunne, Donal Cahill, Elizabeth Kehoe, and Joanna Connolly. This work was supported by Science Foundation Ireland (SFI08/IN.1/B1916-Corvin). The authors would like to acknowledge data management support from the Trinity Centre for High Performance Computing. The data management system used for this work was BC|SNPmax v. 3.5-121 (Biocomputing Platforms Ltd, Finland).






