INTRODUCTION
Non-typhoidal Salmonella infection is a serious threat to global public health, causing an estimated 93·8 million illnesses and 155 000 deaths worldwide every year [Reference Tauxe1, Reference Majowicz2]. Most (95%) Salmonella infections in humans are attributed to ingestion of contaminated foods [Reference Gomez3]. While usually self-limiting in most individuals, salmonellosis may require antimicrobial drug treatment in infants, the elderly, and immunocompromised individuals. However, antimicrobial resistance has become increasingly common among Salmonella. Because antimicrobial resistance varies widely between and within countries, surveillance is essential to provide data on the magnitude and trends in resistance and to monitor the effect of interventions [4]. The emergence and spread of antimicrobial-resistant Salmonella, particularly multidrug-resistant (MDR) strains, has been reported in many countries [Reference Cailhol5–Reference Threlfall9] and is increasingly of major concern as these strains reduce the therapeutic options in cases of invasive infections and have serious public health implications. Unfortunately, the spectrum of antimicrobial resistance in Salmonella is also increasing and extends to resistance to fluoroquinolones (ciprofloxacin) and third- and fourth-generation cephalosporins (ceftriaxone). As these drugs are the primary choice for the treatment of invasive salmonellosis in humans, the appearance of resistance to such antimicrobials is of particular concern.
Despite an estimated high number of salmonellosis cases in many parts of the world, there is a lack of data on the infection and antimicrobial-resistant Salmonella in China. The Shanghai Center for Disease Control and Prevention (SCDC), participating in the World Health Organization Global Foodborne Infections Network (WHO-GFN) since 2005, has performed isolation, identification and characterization of foodborne pathogens recovered from patients. The present report provides data on serovars, and antimicrobial resistance of Salmonella isolated from patients with diarrhoea in Shanghai from 2006 to 2010.
MATERIALS AND METHODS
Specimen collection
This study was conducted at 24 sentinel hospitals and eight regional SCDC diagnostic laboratories located in five districts of Shanghai (Children's Hospital of Shanghai, and Children's Hospital of Fudan University were added to this project in 2010). Physicians were asked to collect stool samples from patients who presented with ⩾3 loose stools within 1 day and had other symptoms such as fever, vomiting, or abdominal pain [10]. Demographic and clinical information for each case, including age, gender, symptoms, date of illness onset, and date of specimen collection, was collected and electronically transmitted to SCDC.
Bacteriological examination
The clinical laboratories used a standardized procedure to isolate Salmonella from stools [11]. Stool specimens were enriched in tetrathionate Brilliant-Green broth or selenite broth for 6–8 h at 37°C, followed by subcultivation on Hektoen Enteric agar, Salmonella-Shigella agar, or CHROMagar Salmonella agar (Becton Dickinson, USA). The plates were incubated at 37°C for 18–24 h and presumptive colonies were further tested using triple-sugar-iron agar, motility indole urea agar, l-lysine decarboxylase, and l-galactosidase (o-nitrophenyl-l-d-galactopyranoside; ONPG). Isolates were confirmed as Salmonella using API 20E test strips (bioMérieux, France). O and H antigens were characterized by slide agglutination with commercial antiserum (S&A Reagents Laboratory, Thailand), and serovars were assigned according to the Kauffmann–White scheme [12].
Antimicrobial susceptibility testing
Isolates were tested for susceptibility to 16 antimicrobials using the Kirby–Bauer disk diffusion method [13]. The antimicrobials were ampicillin (10 μg), cefotaxime (30 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), ceftazidime (30 μg), gentamicin (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), ceftiofur (30μg), amoxicillin/clavulanate acid (30 μg), trimethoprim/sulfamethoxazole (1·25/23·75 μg), ofloxacin (5 μg), cefepime (5 μg), trimethoprim (5 μg), sulfisoxazole (300 μg), and streptomycin (10 μg). Escherichia coli ATCC 25 922 and ATCC 35218 were used as quality control organisms. Results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines [14].
Statistical analysis
A χ 2 or Fisher's exact test was used for data analysis using SAS v. 9.2 (SAS Institute Inc., USA). A P value of < 0·05 was considered statistically significant.
RESULTS
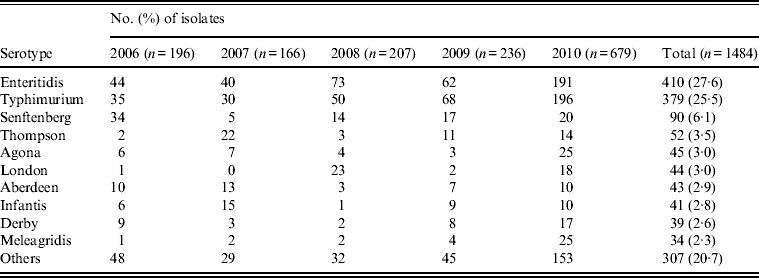
Between January 2006 and December 2010, a total of 18 000 stool specimens were cultured from patients with diarrohea in Shanghai, resulting in 1484 (8·2%) Salmonella isolates. The number of isolates per annum ranged from 166 to 679 over the sample period (Table 1). Seventy serovars were identified with 10 serovars accounting for nearly 80% (1177) of the isolates, chief of which were Enteritidis (410, 27·6%), Typhimurium (379, 25·5%), Senftenberg (90, 6·1%), Thompson (52, 3·5%), and Agona (45, 3·0%). The other 60 serovars were distributed among the remaining 307 isolates (Table 1).
Table 1. Top 10 serovars of Salmonella isolates from humans (2006–2010) in Shanghai, China
The age range of patients spanned from 2 days to 91 years. Of 1204 isolates from 2006 to 2010 (except those from Shanghai Children's Hospital and Children's Hospital of Fudan University), 76·4% were from adults aged 18–60 years. However, most isolates (79·6%) from Shanghai Children's Hospital and Children's Hospital of Fudan University in 2010 were recovered from children aged <4 years (Table 2). The male/female ratio of positive isolation was 1·1:1.
Table 2. Distribution of human Salmonella isolates by gender and age (2006–2010), Shanghai, China
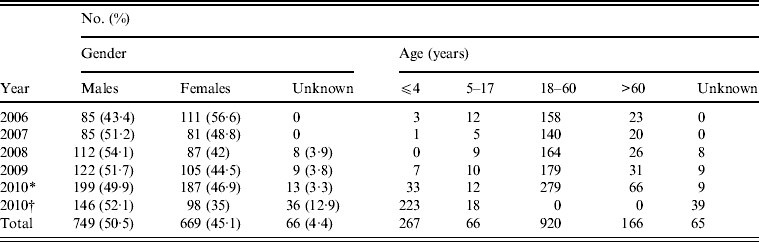
* Salmonella data from Shanghai excluding those from Children's Hospital of Shanghai, and Children's Hospital of Fudan University in 2010.
† Salmonella data from Children's Hospital of Shanghai, and Children's Hospital of Fudan University in 2010.
Most isolates (77·6%) were resistant to one or more antimicrobials and 50·9% were resistant to nalidixic acid, followed by sulfisoxazole (47·9%), streptomycin (37·6%), ampicillin (31·3%), tetracycline (30·5%) (Table 3). Resistance was also observed, but to a lesser extent, to ceftiofur (6·4%), amoxicillin/clavulanate acid (4·7%), ceftazidime (3·8%), cefotaxime (3·6%), ciprofloxacin (2·8%), cefepime (2·1%), and ofloxacin (1·4%). Of the top three serovars (Enteritidis, Typhimurium, Senftenberg), Typhimurium exhibited the highest percentages of resistance to each of these drugs (2·4–6·6%) (Fig. 1), whereas Senftenberg displayed resistance only to ceftazidime (3%).
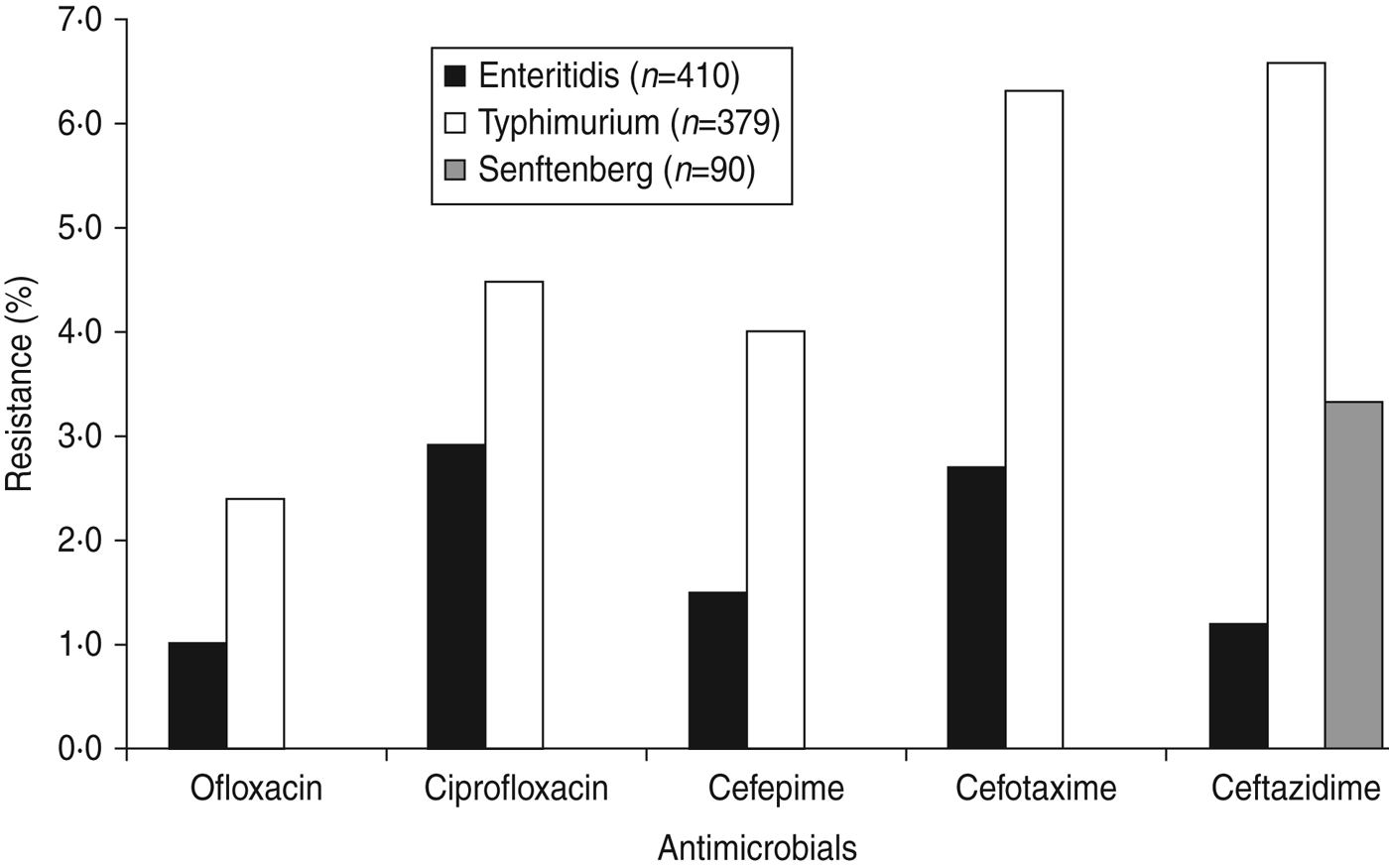
Fig. 1. Resistance to fluoroquinolones, and to third- and fourth-generation cephalosporins in Salmonella serovars Enteritidis, Typhimurium and Senftenberg from humans in Shanghai, China.
Table 3. Antimicrobial resistance of Salmonella isolates from humans in Shanghai, China, 2006–2010
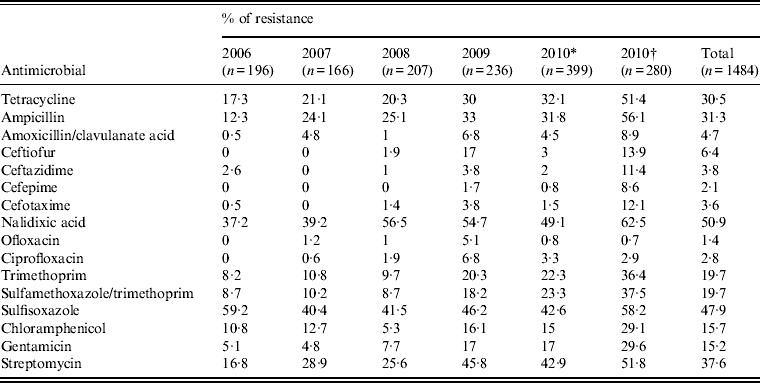
* Antimicrobial resistance data of Salmonella from Shanghai excluding those from Children's Hospital of Shanghai, and Children's Hospital of Fudan University in 2010.
† Antimicrobial resistance data of Salmonella from Children's Hospital of Shanghai, and Children's Hospital of Fudan University in 2010.
The emergence or increase in resistance to certain antimicrobials during the study period is noteworthy (Table 3). All isolates from 2006 were susceptible to ceftiofur, cefepime, ofloxacin, and ciprofloxacin but by 2009, resistance rates of 17% to ceftiofur, 1·7% to cefepime, 5·1% to ofloxacin, and 6·8% to ciprofloxacin were found. Significant increase in resistance to other antimicrobials was also recorded for isolates from 2006 to 2009: tetracycline (17·3–32·1%, P < 0·01), ampicillin (12·3–33%, P < 0·01), amoxicillin/clavulanate acid (0·5–6·8%, P < 0·01), nalidixic acid (37·2–54·7% P < 0·01), trimethoprim (8·2–20·3%, P < 0·01), sulfamethoxazole/trimethoprim (8·7–18·2%, P < 0·01), gentamicin (5·1–17%, P < 0·01), and streptomycin (16·8–45·8%, P < 0·01). Interestingly, most antimicrobial resistance rates (except ciprofloxacin and ofloxacin) of strains isolated from Shanghai Children's Hospital and Children's Hospital of Fudan University were higher than those from other hospitals in 2010 (Table 3), indicating difference in antimicrobial treatment between infections from children and adults. For example, ciprofloxacin is generally not used for young children.
Importantly, a large proportion of the resistant isolates (40·3%) were resistant to ⩾3 antimicrobial agents and these were predominantly the serovars Enteritidis (34·4%) and Typhimurium (33·6%), with the most common resistance phenotypes being nalidixic acid, sulfisoxazole and streptomycin. Thirty-eight isolates were resistant to >10 antimicrobials tested and four isolates to 15 antimicrobials, including sulfonamides, fluoroquinolones, tetracyclines, aminoglycosides and β-lactamases. Eight isolates (three Enteritidis and five Typhimurium) were co-resistant to fluoroquinolones and third- and fourth-generation cephalosporins in addition to 5–10 other antimicrobials.
DISCUSSION
This study provided comprehensive data on serovars and antimicrobial susceptibility of non-typhoidal Salmonella from diarrhoea patients in Shanghai from 2006 to 2010. Enteritidis and Typhimurium were the most common serovars as reported in other parts of the world [Reference Herikstad15, Reference Galanis16], although our other top ranking serovars differed from some countries. For example, Newport, Javiana, I4, [Reference Cailhol5], 12:i:-, and Heidelberg were among the top 10 serovars reported from the USA and Canada. On the other hand, Senftenberg, Thompson, Agona, London, Aberdeen, Derby and Meleagridis were among the top 10 serovars in this series with Senftenberg the third most frequently isolated serovar in Shanghai. Interestingly, this serovar has mostly been described as an uncommon human pathogen in the past but was one of the 10 most prevalent serovars from animal farms in England and Wales [Reference Liebana17], and a predominant serovar from marine environments and seafood, especially in temperate and tropical zones [Reference Martinez-Urtaza18].
The worldwide emergence of resistance to antimicrobials in bacterial pathogens is an important public health problem [Reference Cailhol5, Reference Xia8, Reference Cui19] and an increased number of Salmonella isolates have become multidrug resistant in many developing and developed countries [Reference Ashtiani20]. Resistance to several clinically important drugs including quinolones/fluoroquinolones and the third- and fourth-generation cephalosporins is particularly troublesome. Studies have shown that Salmonella resistant to nalidixic acid also displayed decreased susceptibility to ciprofloxacin, and that infections with ciprofloxacin-resistant Salmonella were associated with increased mortality [Reference Travers and Barza21]. Nalidixic acid resistance in non-typhoid Salmonella was reported in many countries in the mid-1990s [Reference Breuil22–Reference Threlfall24] and accounted for less than 18% of isolates in Europe in 2000 [Reference Threlfall9]. More recently nalidixic acid resistance rates of Salmonella in India and Taiwan (Typhimurium) were reported to be 20% and 21·6%, respectively [Reference Singh25, Reference Torpdahl26], but this contrasts with 2% for all Salmonella and 0·2% which were ciprofloxacin resistant in the USA in 2010, and 3·8% of S. Typhimurium in Korea reported in 2004; none of which were ciprofloxacin resistant [27, Reference Yoo28]. Our finding that nalidixic acid resistance was the most common resistance phenotype (50·9%) in clinical isolates of Salmonella in Shanghai, and that 2·8% were also resistant to ciprofloxacin is therefore noteworthy although similar resistance rates for these drugs have been reported in other regions of China, indicating a national trend [Reference Deng29, Reference Ran30]. The emergence of ciprofloxacin resistance in Salmonella is of public health significance as it indicates possible over-prescription in humans and misuse in animals. This may lead to potential failure in the treatment of patients infected with fluoroquinolone-resistant strains and therefore physicians in Shanghai and China in general need to consider determining the antibiogram of all isolates to ensure effective treatment of patients with Salmonella infection.
Resistance to sulphonamides has been described in the literature to varying degrees in different countries. In the USA, about 28% S. Typhimurium were resistant to sulfisoxazole [27], whereas 97% and 43% of S. Typhimurium were resistant to the drug in Japan and Denmark, respectively [Reference Torpdahl26, Reference Shahada31]. A relatively high level of resistance to sulfisoxazole (47·9%) was also observed in this study.
Cephalosporins are among the most diverse classes of antimicrobials, and are grouped into ‘generations’ by their antimicrobial properties. Each generation has a broader spectrum of activity than a previous one. Resistance to these drugs has been reported in several countries, albeit at a relatively low rate [Reference Torpdahl26, 27]. Unfortunately, the present study revealed the emergence of co-resistance to fluoroquinolones (ciprofloxacin and ofloxacin), and third-generation (ceftazidime and cefotaxime) and fourth-generation (cefepime) cephalosporins in Enteritidis and Typhimurium isolates. These drugs are among the most widely used worldwide, and are considered critically important in human medicine according to WHO criteria [Reference Paterson and Bonomo32] as they are reserved for use in the hospital setting to treat patients with serious and life-threatening infections. The emergence and increased resistance rates to these drugs is therefore particularly alarming. Our findings support the need of continuing active surveillance of antimicrobial resistance in Salmonella and other bacterial pathogens as it is essential to the safe and effective use of antimicrobial drugs for humans thereby protecting public health.
ACKNOWLEDGEMENTS
This work was funded in part by Mega-projects of Science and Technology Research of China (No. 2012ZX10004215–003), the Subproject 6 of the Sino-US EID (Emerging Infectious Diseases) Cooperation Project and the China Postdoctoral Science Foundation funded project (Nos. 2012T50419, 2011M500781)
DECLARATION OF INTEREST
None.






