INTRODUCTION
Campylobacteriosis is New Zealand's most frequently notified bacterial disease with an annualized incidence rate that exceeded 400 cases/100 000 during 2007 [Reference Sears1]. Interventions primarily by the poultry industry have reduced the incidence, and by the end of 2010 the rate was 180 cases/100 000 [2]. While such a marked reduction is impressive, this rate still exceeds that reported in other countries such as Australia (rate of <120 cases/100 000), the UK (rates of 50–107 cases/100 000), and the USA (rates of 10–30 cases/100 000) [Reference Lake3]. Difficulties in attributing sources of infection and uncertainty about the reasons for the high notification rate in New Zealand have hampered risk management. Concern about the rate of campylobacteriosis in New Zealand in the early 1990s prompted researchers to undertake two significant case-control studies [Reference Ikram4, Reference Eberhart-Phillips5]. The major finding of both studies was the significant association of campylobacteriosis with consumption of undercooked poultry, barbecued chicken and poultry eaten away from home. While the combined population attributable risk for chicken-related variables exceeded 50%, a range of other significant risk factors was also identified including contact with an ill person, overseas travel, consumption of unpasteurized dairy products, non-city water outside of the home, rainwater as a source of home water, handling calf or puppy faeces, ownership of puppies or caged birds, and occupational contact with cattle or cattle/calf carcasses [Reference Eberhart-Phillips5]. While these risks are biologically plausible, they were each reported by relatively small numbers of patients, and in many instances, patients reported multiple risk factors making it difficult to determine the actual source of infection for individual patients. Compounding this is the ubiquity of Campylobacter spp. in a wide range of sources. Previous studies in New Zealand have found Campylobacter in 23–87% of raw chicken samples [Reference Lake3, Reference Hudson6, Reference Mullner7], 3–9% of beef, lamb and pork samples [Reference Lake3], 12–90% of animal faecal samples (including cows, sheep, pigs) [Reference Devane8], 12·5% of wildfowl faecal samples [Reference French9], 55–85% of river water samples [Reference Devane8] and infrequently in other foods [Reference Edmonds and Hawke10]. Poultry is clearly an important risk factor in campylobacteriosis and is the most frequently contaminated retail food. Intentional and unintentional interventions in Iceland [Reference Stern11] and Belgium [Reference Vellinga and Van Loock12], respectively, suggest 40–60% of campylobacteriosis cases are related to a poultry source. Recent attribution studies in New Zealand suggest that the original source of isolates causing human disease is chicken in 55–71% of cases, sheep in 15–16% of cases, and cattle in 8–14% of cases [Reference Mullner13, Reference Mullner14]. However, the vehicle or mechanism of infection remains undefined and the effectiveness of any intervention, particularly for non-poultry sources, difficult to measure.
The study of outbreaks is an important tool for understanding and managing many infectious diseases. Indeed many of the risks identified for campylobacteriosis in case-control studies have been supported by outbreak investigations with reports of outbreaks due to contaminated water [Reference Stehr-Green15, Reference Vogt16], an infected food handler [Reference Fitzgerald17], and the consumption of raw milk [Reference Evans18, Reference Harrington19], chicken [Reference Pearson20], chicken liver pâté [Reference Whyte21], and sausages [Reference Graham22]. However, these account for only a small number of cases, and are generally thought to represent unusual events, rather than being representative of most cases. A review of notified campylobacteriosis cases in New Zealand over a 5-year period (2000–2004) identified 216 reported outbreaks involving 992 patients, or 1·7% of the >58 000 notified campylobacteriosis cases in the period examined [Reference Wilson23]. A ‘definite’ source of campylobacteriosis was identified in only five of these outbreaks. Evidence for a vehicle or source of these reported campylobacteriosis outbreaks was derived primarily from a ‘history of exposure to an implicated source’. Only 3% of outbreaks had laboratory evidence on the source, and only 2% involved an epidemiological study. Thus, consistent with previous studies [Reference Devane8], an established dogma of public health has been that campylobacteriosis is a sporadic disease, of which few cases are outbreak related, and genotyping of clinical isolates is of little value. The result is that campylobacteriosis cases are most often not investigated, which perpetuates the problem.
This dogma of sporadic campylobacteriosis has been challenged by a small pilot study in which we genotyped 183 notified human isolates by pulsed-field gel electrophoresis (PFGE) using both SmaI and KpnI [Reference Gilpin24]. Approximately two-thirds of the isolates could be grouped into clusters of between two and 26 isolates with indistinguishable SmaI and KpnI patterns. Clustering of subtypes of human campylobacteriosis isolates in New Zealand was also seen in a study of 112 campylobacteriosis cases during June and July 2006 [Reference McTavish25]. Even though these isolates originated from eight district health board (DHB) regions across New Zealand clustering of PFGE genotypes was still identified among isolates from different regions as well as among isolates from the same region. Seventeen PFGE groups of two isolates or more (81 isolates) were identified, and of these 17 groups, 15 had isolates from two or more DHBs [Reference McTavish25]. However, in these studies, the associated epidemiological information was not detailed enough to allow identification of actual common sources.
In this study we used PFGE to analyse Campylobacter spp. isolated from human cases in the Canterbury region of New Zealand from March 2009 to February 2010. Questionnaires were administered to patients to identify possible common risk factors. To capture a snapshot of genotypes in the poultry and ruminant populations during the study period retail poultry and ruminant offal from a range of supermarket butcheries was also tested for the presence of Campylobacter spp. Using a combined molecular epidemiology approach, a source of campylobacteriosis was assigned for each patient.
METHODS
Campylobacter isolates were obtained from human faecal samples submitted to clinical laboratories in Christchurch and Timaru in Canterbury between April 2009 (week 15) and February 2010 (week 61). The case definition was clinically confirmed cases of campylobacteriosis from the Canterbury region of New Zealand with onset dates between February 2009 and February 2010. In the Canterbury region (an area of 45 346 km2 with a population of 565 800), 70% of people live in Christchurch city and 8% live in Timaru city. The remaining population spans a range of urban to very isolated rural areas. The isolates were obtained from faecal samples streaked onto charcoal-cefoperazone-deoxycholate agar that were incubated microaerobically at 37 °C for 48 h. Colonies suggestive of Campylobacter were confirmed as Gram-negative-curved bacilli by Gram stain. Isolates were stored on charcoal swabs for transport to the central laboratory, where they were re-streaked on Columbia sheep blood agar.
Retail offal samples (primarily livers, hearts and kidneys, n=925) purchased from different supermarkets throughout Christchurch were tested for the presence of Campylobacter spp. Samples were collected over three periods: March–June 2009 (weeks 11–28), November–December 2009 (weeks 45–51), and January–February 2010 (weeks 54–61). One chicken offal sample and one sheep, beef or pork offal sample were collected from each supermarket in any one week. Offal was not imported. Offal (2·5 g) was added to 100 ml of m-Exeter selective enrichment broth, macerated in a stomacher for 1 min and then incubated at 37 °C for 48 h as previously described [Reference Devane8]. The enrichment broth was then plated onto m-Exeter agar, and then Columbia sheep blood agar, before putative Campylobacter isolates were analysed further. The species of each isolates was identified using a multiplex polymerase chain reaction (PCR) assay that detects the presence of Campylobacter jejuni and Campylobacter coli [Reference Wong26], and a real-time PCR assay for detection of Campylobacter lari [Reference Chaban27]. All isolates were analysed by PFGE using the standardized PulseNet protocol [Reference Ribot28], with the Salmonella Braenderup H9812 strain restricted with XbaI as a size standard [Reference Hunter29]. Gels were made with 1% (w/v) SeaKem Gold agarose and electrophoresed for 18 h using an initial switch time of 6·8 s and a final switch time of 38·4 s for SmaI and an initial switch time of 5·2 s and a final switch time of 42·3 s for KpnI. PFGE profiles were analysed and compared using BioNumerics version 5.1 (Applied Maths, Belgium). SmaI and KpnI pattern designations were assigned using a code of Sm or Kp followed by a four-digit number. Isolates with the same pattern number are visibly indistinguishable using an optimization of 0·5% and tolerance of 0·5%. Isolates were also compared with previous New Zealand isolates in a database of 410 human, 287 chicken and 230 ruminant isolates. Selected isolates were analysed by multi-locus sequence typing (MLST) as previously described [Reference McTavish30]. Simpson's diversity index was calculated as 1−∑[n i(n i – 1)/N(N – 1)], where N is number of samples, and n i is the number of samples with genotype i, with the confidence intervals (CI) calculated as previously described [Reference Grundmann, Hori and Tanner31].
Between April and mid-July 2009, patients were contacted by telephone and a questionnaire was administered to explore possible sources of infection and risk factors. Between July and October 2009, a shortened postal questionnaire was sent to patients, and between November 2009 and February 2010 attempts were made to administer a full telephone questionnaire to patients. If this was not successful, a postal questionnaire was sent. A full comparison of questionnaires will be the subject of a forthcoming paper. While all of the questions in the postal questionnaires were included in the telephone questionnaires, the reverse was not true. Consequently for each question the number of responses differed, due to the questionnaire used and the response rates.
Cases were first grouped on the basis of three or more cases with indistinguishable genotypes, which were further examined on the basis of temporal and spatial clustering. Temporal clustering (three or more cases with <4 weeks between isolation dates) of indistinguishable genotypes was assumed to indicate a source common to cases, while genotypes with no temporal clustering were assumed to be more likely the result of individual direct contact with a source or exposure potentially unique to that person. For each genotype group, comparisons were made with genotypes of offal isolates, and self-reported risk factors and exposures examined. For each case a likely source of infection was assigned which were categorized broadly as overseas travel, direct animal contact, foodborne, water exposure, person-to-person and unknown. Cases were assigned to the same source when: (a) cases had a consistent temporal clustering pattern; (b) the same genotype predominated in either a poultry or ruminant offal source and; (c) risk factors were common (or absent) in most cases in that genotype group.
The spatial location of each notified human case was assigned at the meshblock level (small geographical unit of ∼100 people defined by Statistics New Zealand) and a smoothed relative risk surface of campylobacteriosis was determined using a Bayesian model-based approach as previously described [Reference Mullner32]. Briefly, the expected number of cases at time t in meshblock i are modelled using a Poisson(nλ it) distribution, where the rate λit is modelled using multiplicative temporal and spatial random effects. Structural priors on the random effects provide a smoothing in both space and time, allowing the trend in risk to be assessed. In particular, the spatial random effects provide the relative risk in each meshblock.
RESULTS
Spatial distribution
Overall 54% of patients (95% CI 50–58) lived in Christchurch city. There was a higher risk of campylobacteriosis in sparsely populated rural areas than in urban areas (Supplementary Fig. S1).
Animal/food isolates
Of the 923 retail offal samples tested, 484 were positive for Campylobacter spp. (Table 1). Isolates were divided into two categories, of which 240 were poultry isolates (from chicken, turkey and duck offal), and 244 were ruminant isolates (from sheep and beef offal). Campylobacter spp. were not isolated from any of the pork offal samples tested. Species identification of Campylobacter isolates found that 98·1% were C. jejuni, 1·3% were C. coli (three sheep, two turkey, one chicken offal), and 0·6% were C. lari (one each of cow, sheep and chicken offal). SmaI and KpnI PFGE analysis identified 114 different genotypes with a Simpson's diversity index of 0·97 (95% CI 0·96–0·97). Of the 114 genotypes, 49 were only found in poultry offal (112 isolates), 52 were only found in ruminant offal (181 isolates), and 13 were found in both poultry and ruminant offal (129 poultry, 63 ruminant isolates). Among these 13, six were poultry dominant (average 11 poultry isolates for each ruminant isolate), two ruminant dominant (six ruminant isolates for each poultry isolate), while the other five were mixed by source. Compared with isolates previously entered into the database, 72% of the poultry isolates and 51% of the ruminant isolates were new.
Table 1. Prevalence of Campylobacter spp. in offal samples
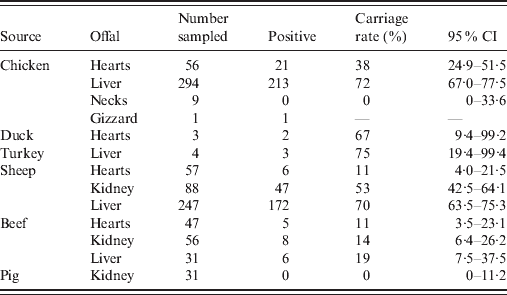
CI, Confidence interval.
Human cases
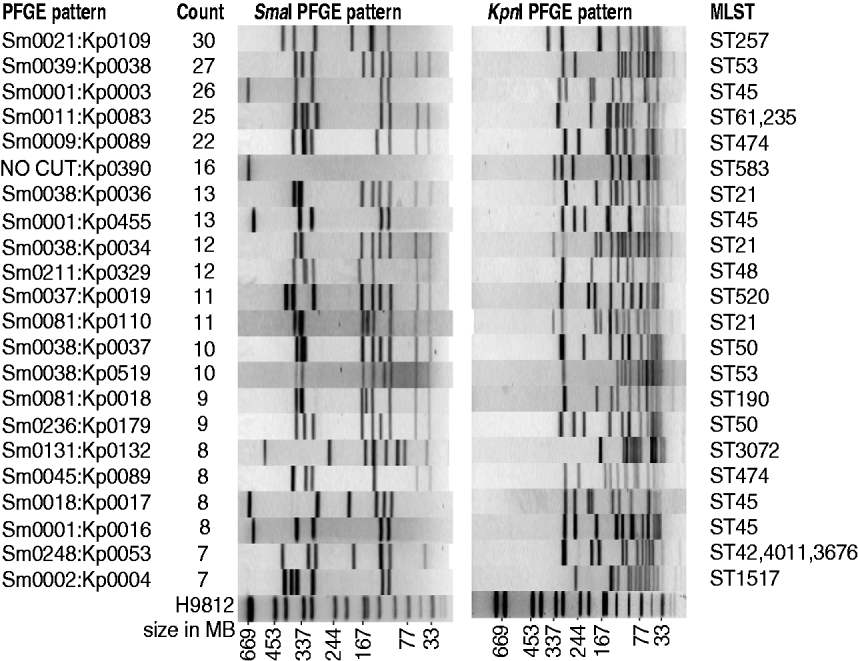
Campylobacter spp. were isolated from 603 human cases of campylobacteriosis during the period March 2009 to February 2010. Of these, 370 (61%) were male and 233 (39%) female. Compared with the general population the 0–4 years and 20–24 years age groups were over-represented in disease incidence rates, while the 10–14 and 30–34 years age groups had lower disease incidence rates (P<0·01). Detailed questionnaires were completed for 364 of these patients, with limited information available about the remaining patients – notably age, sex, occupation and address. Of the 603 human isolates, 572 were identified as C. jejuni, 27 C. coli and four C. lari. The 603 isolates from human cases could be divided into 199 different PFGE SmaI:KpnI genotypes with a Simpson diversity index of 0·985 (95% CI 0·982–0·987). The genotypes are not equally represented among cases, with the 10 most common genotypes accounting for one-third of the isolates, and 22 genotypes accounting for half of the isolates (Fig. 1), while at the other extreme, 20% of isolates, had genotypes that were only observed once (120 genotypes). In comparison with isolates we have genotyped in previous studies (410 human isolates and 633 isolates from other sources), two-thirds of the human isolates had new genotypes.

Fig. 1. The 22 most frequent human PFGE patterns accounting for 50% of all human isolates. Shown are SmaI pattern, KpnI pattern, MLST, and the number of human isolates with that pattern.
Comparison of genotypes found in humans and food
When compared with the 603 human isolates in this study 25 of the poultry or poultry-dominated genotypes (149 offal isolates), were also found in 170 human isolates, and 34 of the ruminant only, or ruminant dominant genotypes (197 offal isolates) were also found among 179 human cases. Of the genotypes that were not observed in any human cases, 25 genotypes were from poultry offal only, 18 from ruminant offal only, and two from both sources.
Identifying the source/pathway for human infection
The process used to assign causes of infection is partially illustrated in Figure 2, with over 70% of cases sharing indistinguishable genotypes with at least two other cases. Of these cases at least 65% displayed some temporal clustering. A likely source of infection was then assigned to 88% of cases by considering spatial and epidemiological information in conjunction with genotype comparisons with isolates from chicken offal and ruminant offal.

Fig. 2 [colour online]. Schematic of strategy used to assign sources of infection, and the percentage of all case sources assigned by each strategy. Common genotypes are ⩾3 human isolates indistinguishable by PFGE using both SmaI and KpnI. Isolates were then classified as having temporal clustering when ⩾3 cases occurred within <4 weeks between isolation dates. Genotypes were then classified as chicken or ruminant dominant, mixed (both chicken and ruminant), or as human only. Spatial clustering was considered along with assessment of reported risk factors and exposures.
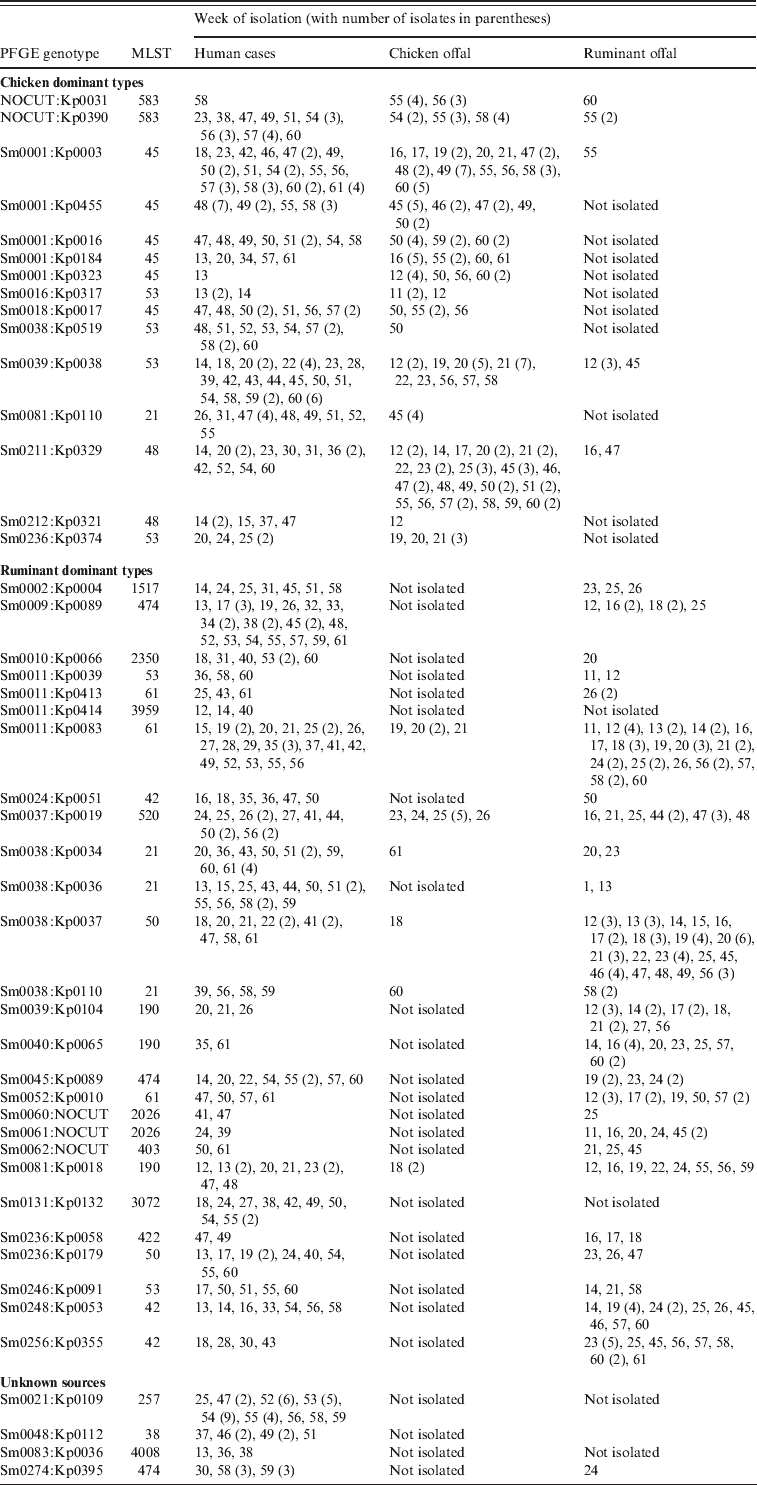
A number of genotypes were associated with a poultry source, often with temporal concordance, the largest groups of which are shown in Table 2. The most common SmaI pattern, Sm0001 was assigned to 71 cases during the study, albeit with 19 different KpnI patterns. Among the offal samples, 75 chicken offal isolates were Sm0001, while just one ruminant offal isolate had this pattern. Those Sm0001 isolates that have MLST were all ST45s. The addition of KpnI typing results allowed the identification of human isolates that cluster temporally with chicken offal isolates for at least five of the Sm0001 genotypes (Kp0003, 16, 184, 323, 455), accounting for 50 of the cases (Table 2). Of these 50 cases, 89% lived in urban areas, and few had contact with ruminant animals. For most an obvious source was not clear, with no common restaurants or specific foods identified. A number of patients did, however, self-report consumption of various chicken dishes, including one who ate a piece of raw chicken before it was cooked.
Table 2. Weeks when specific genotypes of Campylobacter were isolated from human cases, chicken offal and ruminant offal. All genotypes were C. jejuni except for Sm0131:Kp0132 which was C. coli

Among the cases with Sm0001 patterns that did not form a temporal cluster, five were attributed to live chicken contact (including one who ate chicken faeces), and one overseas travel (unique type). Four were also attributed to person-to-person transmission, although the typing would suggest the index case was infected from a poultry source.
Some genotypes such as Sm0236:Kp0374 were only found in chicken offal over a short time period (in this case weeks 19–21), which preceded the observation of this type in four human cases during weeks 20–25. These four cases had no farm animal contact, no overseas travel, no water risks, one had chickens at home and all four reported various chicken foods as suspected sources. Other genotypes such as Sm0211:Kp0329 and Sm0039:Kp0038 were found in chicken offal throughout the year, and were also found in human cases during the year (Table 2), commonly in separate temporal clusters. Eight of the 12 Sm0211:Kp0329 cases had good epidemiological information. None reported overseas travel, water risks or contact with chickens or farm animals. All eight, however, reported various chicken-based foods as suspected sources. A similar set of exposures were observed for the Sm0039:Kp0038 cases with one patient reporting consumption of a raw chicken enchilada. The very strong evidence from one case strengthened confidence in assigning a poultry source of infection for the other temporally clustered cases with the same genotype. Of the chicken-associated cases 71% lived in Christchurch (95% CI 63–78).
While most chicken-associated cases were foodborne, contact with live chickens was reported by 9·2% of cases, with genotyping obtained from 34 cases. Seven cases formed temporal clusters with other cases who did not report animal contact, which when analysed suggested a foodborne chicken source. The remaining 27 were all attributed to direct animal contact, of which contact with live chickens was the only or most likely risk for 17 cases. Occupational exposure to ruminants was the likely source for five of the remaining 10 cases (two dairy farmers, a vet, an artificial inseminator and a slaughterhouse worker). The remaining five cases had multiple direct animal exposures including to cows, sheep, ducks and chickens.
A number of other human cases had Campylobacter genotypes which were not observed in chicken offal, but which were indistinguishable from isolates found in ruminant offal (Table 2). For example, Sm0002:Kp0004 was isolated from seven cases with no clear spatial or temporal clustering. Two patients were farmers, one patient was a slaughterhouse worker, one patient reported contact with cows and consumed unpasteurized milk, one patient suspected person-to-person transmission and the other two patients had limited epidemiological information, although one lived in a rural area and the other was a child aged <5 years. There were no other significant risk factors reported for these patients.
A second scenario among ruminant-associated genotypes was demonstrated by Sm0038:Kp0037 which was isolated from ruminant offal throughout the year. A temporal cluster of five cases within 4 weeks was observed, and patients with this genotype also formed a loose spatial cluster with seven of 10 patients living in Christchurch. None had any farm animal contact, most reported eating at various restaurants with one eating lamb's liver at a restaurant and two drank stream water. However, the stream water contact cases clustered with other cases who did not report this exposure suggesting instead a different common source. On balance, cases were assigned a food, but not a chicken source. Of the ruminant-associated patients, 43% lived in Christchurch (95% CI 36–51).
While Campylobacter genotypes may predominantly be found in certain hosts, our data suggest that most can be found in multiple hosts. For example Sm0037:Kp0019 was found in ruminant offal through the year, and it was found in chicken offal over a 3-week period and at the same time that five cases were notified with campylobacteriosis (Table 2). These five patients lived in different parts of Christchurch, with one on the outskirts. None had contact with farm animals, although one had contact with sick possums, which in previous work have not been found to carry C. jejuni [Reference Devane8]. These five patients therefore, were assigned to a chicken foodborne source. Another six cases of this genotype occurred later in the study, from weeks 41 to 57. These patients were distinctly more rural, with three being dairy farmers, one a shearer, and one a slaughterhouse worker. For these patients, direct contact with ruminant animals appeared to be the most likely scenario for infection.
The most common single genotype recovered from human cases was Sm211:Kp329 (Table 2). The first patient with this genotype was observed in week 25, and was a dairy farmer for whom occupational exposure to cow faeces was the most likely source. For the next 22 weeks this genotype was not observed, before a cluster of 29 cases was observed, 24 of these over a 4-week period. This genotype was not isolated from any offal samples during the study. Previous typing has recovered this genotype from chicken meat and ruminant faeces. All of the patients expressing this genotype were aged between 17 and 80 years, and 80% lived in urban areas (although widely spread in both Christchurch and Timaru). Only 15 had detailed epidemiological data (the Christmas holiday period reduced the response rate), of which one reported ruminant animal contact, none had contact with chickens, two had water exposures and one a barbeque involving chicken. No common food premises were evident. This was clearly a common-source outbreak, for which a source could not be definitively identified. However, a number of sources could be excluded – namely overseas travel, direct animal contact, drinking water, and recreational water exposure. The only plausible remaining exposure was food-related exposure from a widely distributed food which could have been a poultry or non-poultry source.
Overseas travel was reported by 29 patients, with the genotypes of 20 of these cases not previously isolated from any animal or food sources in New Zealand. Of the other nine cases eight had genotypes that had been isolated from chicken sources and one genotype had been isolated from a ruminant source. Of the human-only isolates, 13 were only observed once in this study. Of the other seven human-only isolates, two were a husband and wife who had been in Australia, while another two had visited Bali and Australia, but 5 months apart. The other three cases had the same genotype as three other cases who were not questioned. Apart from the husband and wife, no temporal or spatial clustering was observed in these cases. There was strong molecular epidemiological evidence for 25 of these patients becoming infected overseas. The other four patients were overseas for only part of the possible incubation period. Two had genotypes which formed temporal clusters with other human patients who did not report travel and the source of infection for these two patients was most likely a common source food outbreak (chicken). One patient reporting overseas travel was a stock truck driver with a ruminant Campylobacter genotype who was uncertain when the illness started. For this patient the source was assigned as direct animal contact (occupational exposure). The final patient reported overseas travel at the edge of the incubation period (10 days before onset). The genotype of this case was indistinguishable from one isolated from chicken offal 2 months earlier. The patient also reported water contact, contact with chicken faeces and worked in a restaurant. Multiple unresolved risk factors resulted in the source of infection being assigned as unknown for this patient.
Definitive evidence for waterborne transmission was difficult to achieve due to multiple exposures of many patients reporting potential waterborne risks. Drinking water was assigned as the most likely source for five patients whose only risk factor was the consumption of river water and other high-risk water sources. Another seven patients were assigned to recreational water exposure where this was their only reported risk factor. The genotypes of these cases were all ruminant or from unknown sources.
Eighteen percent of patients reported contact with at least one other person who was sick at the same time, or in the 10 days before they became sick. This was assigned as the most likely source of infection for 16 patients. A number of these were within-family transmissions, often where the initial case was not notified. Person-to-person transmission may actually be higher than reported in this study because when cases were part of a common genotype ‘outbreak’ they were usually assigned to that source, even though they may have been a secondary case.
Of the 603 human isolates, 27 were C. coli, and they could be divided in 16 genotypes. Thirteen genotypes were only observed once, while the most common genotype (Sm0131:Kp0132) was recovered eight times (Table 2). Four of the C. coli cases were acquired overseas. Of the locally acquired cases direct animal contact was the likely source for 12 of these including three slaughterhouse workers, one stock truck driver, three patients with cow contact, four patients with sheep contact, and one patient reporting consumption of pork from a wild pig. Only five of the 23 locally acquired patients lived in Christchurch.
Four of the 603 human isolates were C. lari. All were rural patients, with no spatial or temporal clustering of these isolates, despite two having the same genotype. One patient was a slaughterhouse worker who slaughtered deer, the second patient had exposure to recreational water, and the third patient had contact with recreational water, domestic chickens and roof water. The fourth patient had insufficient epidemiological data from which to determine a likely source.
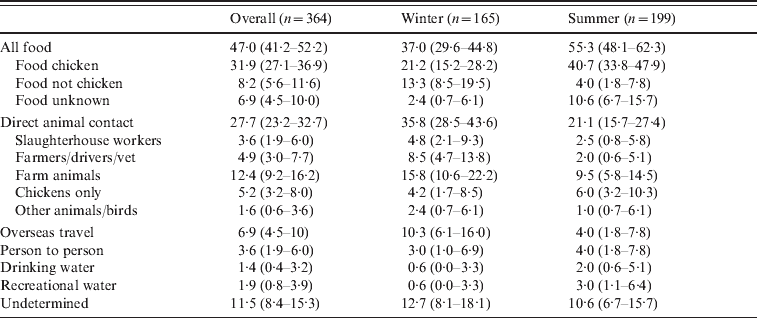
Table 3 summarizes the overall assignment of sources based on the 364 patients with both genotyping and detailed epidemiology. Overall the source of infection was food for 47·0% of patients, direct animal contact for 27·7% of patients, overseas travel for 6·9% of patients, person-to-person contact for 3·6% of patients, water for 3·3% of patients, and for 11·5% of patients a source could not be determined. Of cases attributed to a food source, 68% were attributed to chicken, 17% to non-chicken sources and for 15% the source could be either chicken or non-chicken. Of these food-related cases, 47% ate at a restaurant or ate takeaways. Direct animal contact was the result of occupational contact (a slaughterhouse worker, farmers, stock truck drivers, and a vet – 8·5% of cases), other direct contact with farm animals (12·4%), contact with domestic chickens (5·2%), and contact with pets, wildfowl or other animals (1·6%). Cases were separated into a winter period (April–October 2009) and a summer period (November 2009–February 2010). During the summer period 55% of cases were attributed to food, while during the winter months 37% of cases were attributed to food. In contrast we attributed 21% of cases during the summer to direct animal contact, as opposed to 36% of cases during the winter months.
Table 3. Sources of infection for cases of campylobacteriosis

Values are % (95% confidence interval).
DISCUSSION
The hypothesis of this study was that by combining genotyping of human Campylobacter isolates, with concurrent genotyping of food and environmental isolates, and detailed questionnaires of patients, the specific causes of campylobacteriosis could be identified. Resource restrictions limited the number and variety of potential food and environmental sources of Campylobacter spp. that were tested. In this study sources were restricted to retail offal samples obtained from 33 supermarkets throughout Christchurch. These were tested to sample the types of Campylobacter spp. circulating in the supermarket poultry and in the ruminant animal population, and not because it was suspected that contact or consumption of offal was a significant source of campylobacteriosis (only two patients reported contact with chicken offal, and eight with ruminant offal). High prevalence rates in chicken and sheep liver in particular facilitated the recovery of Campylobacter isolates for comparison. Previous studies have also shown that the genotypes in offal are similar to those causing disease in humans [Reference Strachan33]. The significant proportions of new genotypes emphasized the importance of genotyping source isolates and when poultry isolates, in particular, were found in retail stores concurrently with the emergence of human cases of the same genotype this provided strong evidence for a poultry source. Sampling of poultry has the potential to bias results towards finding poultry as a source. However, as illustrated in this study, it can also largely exonerate poultry as a source. To truly achieve this, a more systematic and comprehensive testing of poultry isolates is needed, and must always be tempered by the observation that genotypes of Campylobacter can and do infect multiple species.
Following this strategy we assigned a source of infection to 88% of patients in Canterbury between April 2009 and February 2010. New Zealand like most other countries has a pronounced summer peak of cases, with over twice as many cases presenting during the summer period. Our analysis suggests that during this summer period 55% of cases were food related, while during the winter period just 37% were food related. Of these food-related cases, less than half reported buying takeaways or eating at a restaurant. This suggests that at least half of the food-related cases resulted from food prepared at home, and the proportion is likely to be much higher, as no common commercial food premises were identified among cases. While over two-thirds of the food-related cases were attributed to chicken, only six patients reported eating raw or undercooked chicken. One of these was a dairy farmer with a Campylobacter genotype consistent with a ruminant source, suggesting consumption of undercooked chicken was an unlikely source of his campylobacteriosis. Raw or undercooked chicken would appear to account for a maximum of just 1·4% of patients. Cross-contamination of either cooked chicken or other foods by uncooked chicken would appear a more likely mechanism of transmission in food-related cases. This is difficult to produce conclusive evidence for as it relies largely on an absence of other sources and genotyping, but the summer peak of cases linked to chicken suggests efforts targeting reducing Campylobacter carriage by chickens at this time of the year would be beneficial.
For 36% of patients in winter and 21% of patients in summer, the most likely source of campylobacteriosis was direct animal contact. Occupational contact by slaughterhouse workers, farmers, stock truck drivers, and a vet accounted for almost 9% of all cases. Meat processors with genotypes that were part of an outbreak, may have contracted the infection at work, or acquired it subsequently from contaminated food. This is difficult to tease out, and for the few cases where this occurred in the study we attributed the case to an outbreak-related source rather than the potential occupational source. While good hygiene practices can reduce the chances of becoming ill, the high prevalence of Campylobacter and the ubiquity of faecal material in farming, make potential infection opportunities almost unavoidable in the long run. Several of these people reported previous illness, suggesting that immunity in slaughterhouse workers [Reference Cawthraw34] may only be partial.
We attributed 5·2% of all cases to contact with domestic chickens, and this was also a risk factor in another 4·2% of patients. A previous study that focused on a more rural area identified contact with live chickens as a major risk factor [Reference Devane8]. In a separate study of 33 domestic poultry flocks we found Campylobacter spp. to be common with genotypes indistinguishable from those found among human patients and among commercial chicken isolates [Reference Anderson, Horn and Gilpin35]. For one of the patients in this study we sampled their home chickens and recovered the same genotype that caused their illness. We believe this provides good evidence for domestic chicken flocks being a source of Campylobacter, and one where intervention strategies, perhaps with a focus on education, could potentially be targeted.
The proportion of cases attributed to chicken in this study is lower than reported in previous attribution studies based on the prevalence of particular MLST genotypes in various sources. For example, a large study in Scotland attributed 76% of campylobacteriosis cases to chicken sources using MLST [Reference Sheppard36]. However, they did note differences among population subgroups. For example among rural young children <20% of cases were attributed to chicken [Reference Strachan37]. The Manawatu area of the North Island of New Zealand has been the focus of a number of attribution studies, which have until recently also attributed at least 60% of cases to chicken [Reference Mullner13, Reference Mullner14]. Following the decrease in numbers of campylobacteriosis cases observed after interventions in the poultry industry, the most recent attribution calculations are more consistent with those reported here, with less than half of cases now attributed to chicken sources [Reference Sears1].
Typically a species is not determined for most cases of campylobacteriosis. In this study 95% of cases were C. jejuni, with the remainder mostly C. coli and a few C. lari. The non-C. jejuni infections appeared to be primarily separate direct infections from animal or water sources. Even fewer Campylobacter cases are genotyped, which as demonstrated by this study is essential to understand the epidemiology of cases, and to assign likely source of infection. Even with a large cluster of cases such as Sm0021:Kp0109, during the 4 weeks when most of the cases with this genotype were observed (24 cases), there were another 69 cases, with 48 different genotypes during the same period. Without genotyping it would have been impossible to even know this outbreak existed, let alone to focus on it. Offal sampling was stopped during this period, so if poultry was the source, we may have missed it. The genotype Sm0021:Kp0109 has previously been recovered from poultry meat [Reference McTavish25] and prior to that ruminant faeces [Reference Gilpin38], while MLST type 257 was assigned in a previous study to poultry with 98% probability [Reference Mullner32]. We can also exclude occupational contact, direct animal contact and a range of other sources. It is tempting to speculate that seasonal food such as Christmas turkey, or another distributed food may be a source. While it is disappointing not to have identified a more conclusive source, this cluster of cases points to the value of a molecular epidemiological approach. The criteria of a minimum cluster size of three isolates with temporal clustering within 4 weeks may need to be adjusted according to the frequency of the genotype, possible source being investigated, and the nature of food distribution networks. Campylobacteriosis has clear differences between urban and rural populations [Reference Strachan37], which for some countries means targeting the strategies in this paper differently for urban and rural groups may be prudent. PFGE appears to provide an appropriate level of discrimination, but it is a cumbersome technique. Reproducibility issues have largely been eliminated through adherence to standardized protocols, but the time taken to complete the test and its cost have little apparent room for further reduction. A number of other methodologies have been developed or are in development. The ideal method will be applicable to a single colony, and produce easily interpretable results within 24 h. MLST has been successfully used for genotyping of Campylobacter in a number of studies [Reference Mullner7, Reference Mullner32, Reference Carter39]. The sources assigned by PFGE in this study, were consistent with sources assigned by MLST in previous studies. For example, all but one of the chicken-dominant genotypes in Table 3 had MLST types previously identified as poultry dominant [Reference Mullner32]. To fit our desired criteria, MLST would need to include additional loci, and remove the individual PCR steps, perhaps via direct whole-genome sequencing. ‘Laddering’ of multiple sequencing reactions within one run could provide simultaneous sequences of multiple isolates at reduced cost. Another approach could be PCR-based binary typing assays such as P-BIT [Reference Cornelius40]. To be practical, multiplexing of the assays is required, as it could then produce results rapidly within 4–6 h after isolation. The key is that whatever the method used, it must allow high-throughput genotyping of all human isolates, and as many isolates from possible sources concurrently. This will be essential particularly for larger populations if this approach is to be feasible. These results could be available before epidemiological questionnaires are even administered. Investigations could then focus resources on clusters of cases with a historical picture of genotypes enabling the significance of clusters of cases to be evaluated. Some genotypes are clearly more common, and a few cases clustering together temporally may not indicate a common source. However temporal clustering of even three isolates of a ‘new’ genotype may allow a common source to be identified. There are many sources of campylobacteriosis, many only accounting for a few percent of the total cases at any one time, but together they add up to a significant disease burden. This paper demonstrates that the molecular epidemiology approach is well worth pursuing, and that sources of campylobacteriosis at both the individual and the community level can be determined. The effectiveness of interventions, education and other measures can then be determined with more confidence and their success measured.
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268812001719.
ACKNOWLEDGEMENTS
We acknowledge financial support for this project from the New Zealand Health Research Council. We also acknowledge the contributions to this project made by many staff from the Canterbury District Health Board, from clinical laboratories, and from ESR staff in Water, Food and Health Groups. We thank Dr Peter Mitchell, Dr Beverley Horne and Dr Hilary Michie for critical comments on the manuscript.
DECLARATION OF INTEREST
None.