INTRODUCTION
Bacteria of the genus Shigella are the causative agent of bacillary dysentery or shigellosis, of which there are an estimated 165 million cases worldwide each year [Reference Kotloff1]. Shigella are clones of Escherichia coli, with the artificial genus boundary encompassing some but not all of the E. coli strains that have evolved a phenotype characterized by the ability to invade and replicate within the colonic and rectal epithelium of primates and humans. In the case of Shigella flexneri, the traditional species boundary is useful because it describes a discrete set of closely related strains that are biochemically and antigenically similar. S. flexneri accounts for more disease world wide than any other dysentery-causing form of E. coli [Reference Kotloff1].
The basic O-antigen repeat unit in the lipopolysaccharide (LPS) of all S. flexneri is a tetrasaccharide comprised of a single N-acetyl glucosamine and three rhamnose residues [Reference Carlin2]. The 12 traditionally recognized serotypes of S. flexneri differ by the addition of glucosyl and/or O-acetyl groups to different sugars in the tetrasaccharide repeat unit [Reference Allison and Verma3]. The exception to this is serotype 6, which is only distantly related to other S. flexneri strains [Reference Lan4], has a different O-antigen repeat unit [Reference Dmitriev5] and is not considered here to be a member of the species. Antisera directed against the traditionally recognized S. flexneri serotypes is commercially available and is used for serotype identification in most epidemiological studies.
In the late 1980s, a novel S. flexneri serotype was identified in Bangladesh and named serotype 1c due to its similarity with other serotype 1 strains [Reference Wehler and Carlin6]. Serotype 1c contains a disaccharide linked to the N-acetyl glucosamine in the tetrasaccharide of its O-antigen, whereas serotype 1a and 1b strains contain only a monosaccharide at this site (Fig. 1). Currently there is no genetic test or commercially available antibodies for the detection of serotype 1c.

Fig. 1. Chemical structure of the tetrasaccharide repeat units in the O-antigens of S. flexneri serotypes 1a and 1c. d-GlcNAc, N-acetylglucosamine; Rha, rhamnose.
Immunity to S. flexneri is serotype-specific. Hence, vaccine strategies to protect against S. flexneri are reliant on knowledge of the most common serotypes present in a geographical area. Serotype 2a has been the focus of most S. flexneri candidate vaccines because it has been identified as the most common serotype in a wide range of epidemiological studies and is thought to be responsible for about 45% of all S. flexneri disease world wide [Reference Kotloff1]. Previously, Isenbarger et al. [Reference Isenbarger7] investigated the prevalence of different bacterial pathogens responsible for diarrhoea in Vietnamese children in Son Tay Province. They identified 77 S. flexneri strains (excluding serotype 6) based on biochemical properties, but found that more than 40% of these were untypable using commercially available antisera. It was noted that a S. flexneri vaccine targeting serotype 2a would have little impact on the diarrhoea burden in Vietnam as serotype 2a accounted for only 6% of S. flexneri isolates.
In this study, we have characterized the 37 untypable isolates from Isenbarger et al.'s survey. The majority of these isolates belong to the newly recognized S. flexneri serotype 1c. Multi locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) showed that these serotype 1c strains consisted of at least 12 different clones, most of which were related to each other but not to serotype 1c reference strains from Bangladesh.
METHODS
Bacterial strains
The 37 untypable S. flexneri strains from Isenbarger et al.'s study were isolated between August 1998 and July 1999 from children living in three communes (Phu Chao, Phu Phuong, Chao Son) in Son Tay Province, 50 km northwest of Hanoi in northern Vietnam [Reference Isenbarger7]. Additional untypable strains isolated from patients in the same area after 1999 were also available and used for PFGE comparisons. Strain 481NT2 was isolated from the Pasteur Institute in Nha Trang, about 1300 km south of Son Tay Province. The serotype 1c reference strains from Bangladesh were isolated from patients attending the Dhaka treatment centre operated by the International Centre for Diarrhoeal Disease Research, Bangladesh. All strains were grown aerobically in Luria–Bertani (LB) medium at 30°C or on LB agar plates.
Serotyping
Serotyping was performed by slide agglutination. A sterile loop was used to mix bacteria from LB agar plates with a drop of antibody on a glass slide. The slide was gently agitated while observing for agglutination. Negative controls were performed using 0·9% NaCl instead of antibody. Isolates were tested using the monoclonal antibodies MASF B and MASF 1c [Reference Carlin and Lindberg8] and also commercial antisera (Denka Seiken, Tokyo, Japan) directed against Shigella spp. group factor antigens and S. flexneri type and group factor antigens.
DNA techniques
Plasmid DNA was prepared by the alkaline lysis method with SDS [Reference Sambrook, Fritsch and Maniatis9]. Genomic DNA was isolated as described elsewhere [Reference Sambrook, Fritsch and Maniatis9]. Oligonucleotide primers used for PCR were purchased from Invitrogen (Carlsbad, CA, USA) or Sigma Proligo (Australia). PCR was performed using Pfu polymerase (Promega, Madison, WI, USA) according to the manufacturer's instructions. DNA sequencing was performed using the Big Dye Version 3.1 sequencing protocol (Applied Biosystems, Foster City, CA, USA) and analysed with the ABI 3730 capillary sequence analyser (Applied Biosytems) at the Biomolecular Resources Facility, John Curtin School of Medical Research, Australian National University.
MLST
MLST was performed using up to seven E. coli housekeeping genes to allow comparison to strains in the Multi Locus Sequence Typing Database for Pathogenic E. coli Version 1.1 (http://www.shigatox.net/mlst), which is operated by the Microbial Evolution Laboratory at Michigan State University with support from the National Institute of Health. The primers and protocols used are described on the database website. Where sequence type numbers are quoted from the database, they refer to sequence types based on seven genes (st7). In some instances it was possible to assign an isolate to a particular st7 without sequencing all seven genes. For at least one isolate from each st7, all seven genes were sequenced.
LPS profiling
LPS was extracted by the whole-cell lysate method. Briefly, 109 cells from log phase culture were pelleted and resuspended in 100 μl of 2× sample loading buffer [10% SDS (w/v), 20% glycerol (v/v), 0·1% Bromophenol Blue (w/v), 0·5 m Tris–HCl (pH 6·8)]. Fifty micrograms of Proteinase K (Roche Diagnostics, Castle Hill, Australia) was added and the sample incubated at 56°C for 1 h. Samples were stored at −20°C until required. Prior to SDS–polyacrylamide gel electrophoresis (SDS–PAGE), 2-mercaptoethanol (final concentration 5%, v/v) was added and the sample incubated at 95°C for 5 min. SDS–PAGE was performed using 4–20% iGels (Life Therapeutics, Clarkston, GA, USA) and LPS was silver stained as described elsewhere [Reference Hitchcock and Brown10].
Clinical symptoms
Data of clinical symptoms were collected for S. flexneri cases that were included in the study published by Isenbarger et al. [Reference Isenbarger7] and additional cases from the same area and time period.
PFGE
Genomic DNA was prepared in agarose plugs [Reference Heath11] and digested with 10 U of XbaI (Fermentas Life Sciences, Vilinius, Lithuania) overnight at 37°C. PFGE was performed using the Bio-Rad CHEF-DR III system (Bio-Rad Laboratories, Hercules, CA, USA), at 6 V/cm, 14°C, with an included angle of 120°, a run time of 21 h and initial and final switch times of 2 s and 30 s, respectively. Gels were stained with ethidium bromide and analysed using BioNumerics version 4.6 software (Applied Maths, Austin, TX, USA). Band sizes were calculated against the PFGE midrange marker 1 from New England BioLabs (Beverly, MA, USA). Cluster analysis was performed using the unweighted pair-group method using arithmetic averages (UPGMA) [Reference Sokal and Michener12].
RESULTS
Antigenic typing
The untypable isolates from Isenbarger et al.'s [Reference Isenbarger7] study were subjected to agglutination testing using Shigella-specific antisera and monoclonal antibodies (Table 1). Twenty-four of the 37 untypable S. flexneri isolates agglutinated with the monoclonal antibody MASF 1c, demonstrating that they were serotype 1c strains. Twelve isolates did not agglutinate with any antibodies and following silver staining of LPS, were found to lack O-antigen (data not shown). A single isolate (isolate 1419) showed an unusual agglutination pattern, reacting with S. flexneri antisera (although not with any serotype-specific antisera), and also with antisera directed against S. dysenteriae serotypes. Silver staining of LPS showed that O-antigen was expressed in this strain, demonstrating that this unusual agglutination pattern did not arise due to autoagglutination. We also obtained a S. flexneri isolate (481NT2) from the city of Nha Trang, which also agglutinated with MASF 1c.
Table 1. Slide agglutination of ‘untypable’ S. flexneri isolates from Son Tay Province using Shigella-specific antisera and monoclonal antibodies
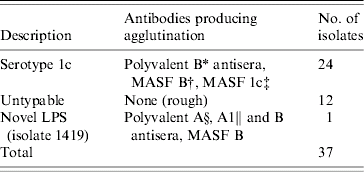
* Antisera specific to S. flexneri and S. flexneri serotype 6.
† Monoclonal antibody specific to S. flexneri, S. flexneri serotype 6 and S. dysenteriae serotype 1.
‡ Monoclonal antibody specfic to S. flexneri serotype 1c.
§ Antisera specific for S. dysenteriae serotypes 1–7.
∥ Antisera specific for S. dysenteriae serotypes 8–12.
MLST
MLST was undertaken to confirm that the rough isolates were in fact S. flexneri, and if so, to indicate which serotypes they may have been derived from. MLST was performed for all of the rough isolates and a selection of serotype 1c isolates (at least one isolate from each plasmid profile type). The 1c isolates and ten of the 12 rough isolates were found to belong to the same sequence type, st7-50. Reference serotype 1c strains from Bangladesh also belonged to st7-50. One of the rough strains belonged to st7-52, which also includes serotype 1 and serotype 2 strains according to the Multi Locus Sequence Typing Database for Pathogenic E. coli Version 1.1. The remaining rough isolate was not a member of the S. flexneri clonal group and belonged to st7-216, which includes S. flexneri serotype 6. Hence, MLST indicated that 10 of the 12 rough isolates may be derived from serotype 1c strains, whereas the remaining two rough isolates were derived from other serotypes.
MLST was also performed for isolate 1419, which had shown the unusual agglutination pattern. Isolate 1419 was found to belong to st7-22 and lay outside the S. flexneri clonal group.
Plasmid profiling
Plasmid profiling was performed for all serotype 1c and rough st7-50 strains in order to compare the plasmid pool to that reported for serotype 1 strains isolated in Bangladesh [Reference Talukder13]. The 140 MDa invasion plasmid was ignored for this analysis as it is frequently lost and was presumably originally present in all isolates for them to have caused disease. Five different plasmid profiles (P1–P5) were observed and are shown in Figure 2. Sixty-three per cent of 1c isolates and 70% of rough st7-50 isolates had the same plasmid profile (profile P5). This same profile was observed in 26% of serotype 1 strains isolated in Bangladesh and examined by Talukder et al. [Reference Talukder13] and also in the serotype 1c strain we had obtained from Nha Trang, Vietnam. Isolates with the plasmid profiles P1, P2 and P3 contained plasmids that corresponded in size to those observed by Talukder et al., although the plasmids were present in novel combinations compared to the isolates from Bangladesh [Reference Talukder13]. One serotype 1c isolate had 2·7 MDa and 2·9 MDa plasmids that were not present in any of the isolates from Bangladesh [Reference Talukder13]. All rough st7-50 isolates had plasmid profiles that were identical to those of confirmed serotype 1c isolates.

Fig. 2. Plasmid profiles for serotype 1c and rough st7-50 isolates. A representative isolate is shown for each of the five plasmid profiles observed (P1–P5). Plasmid sizes (MDa) are listed below each lane and were estimated by comparison to reference serotype 1c strains from Bangladesh [Reference Talukder13]. The plasmids pUC18 (Fermentas Life Sciences) and pBCSK+ (Stratagene, West Cedar Creek, TX, USA) are shown for reference.
PFGE
PFGE is more sensitive than MLST for discriminating between related strains, and was used here to compare all Vietnamese serotype 1c and rough st7-50 isolates that we had available. Following digestion of genomic DNA with XbaI, 12 different PFGE patterns were identified (X1–X12, Fig. 3a) and their relatedness determined using cluster analysis (Fig. 3b). Ten of the clonal patterns were classed as ‘closely related’ or ‘possibly related’ to each other according to the criteria described by Tenover et al. [Reference Tenover14]. Two clonal patterns (X10 and X12) were ‘unrelated’ to any of the other isolates according to Tenover's criteria, although X10 did share some bands in common with the other isolates. The serotype 1c isolate from Nha Trang had the pattern X5, which was the most common pattern observed for serotype 1c isolates from Son Tay Province. All of the rough st7-50 isolates had identical patterns to serotype 1c isolates or patterns that were ‘closely related’ to serotype 1c isolates. We conclude that the rough st7-50 isolates are almost certainly derived from serotype 1c strains. PFGE was also performed for two reference serotype 1c strains isolated in Bangladesh (banding patterns X13 and X14). According to Tenover's criteria, these isolates were unrelated to all Vietnamese serotype 1c isolates.

Fig. 3. (a) PFGE banding patterns for serotype 1c and rough st7-50 clones following digestion of genomic DNA with XbaI. Molecular sizes are shown for reference. Patterns X1–X12 are from strains isolated in Vietnam, while patterns X13 and X14 correspond to reference serotype 1c strains from Bangladesh [Reference Talukder13]. (b) Phylogenetic tree of serotype 1c and rough st7-50 clones, following cluster analysis of PFGE banding patterns. Arrows indicating cut-off levels for degree of relatedness according to Tenover et al.'s [Reference Tenover14] criteria are shown. The number of serotype 1c and rough st7-50 isolates corresponding to each pattern are shown.
Morbidity
We compared the relative occurrence of disease symptoms in patients infected with serotype 1c strains with those recorded for patients infected with other S. flexneri serotypes. We also compared both patient groups with the 10 patients infected with the rough S. flexneri isolates that belonged to st7-50. There was no significant difference in the occurrence of fever, abdominal pain, vomiting, liquid stool, soft stool, bloody stool or mucoid stool between any of the three patient groups (Table 2).
Table 2. Occurrence of clinical symptoms in S. flexneri patients in Son Tay Province

* Per cent of cases where symptom was reported.
† Significance test for a difference in two proportions: P<0·05 was considered significant.
DISCUSSION
Twenty-four of the 37 untypable S. flexneri isolates collected by Isenbarger et al. [Reference Isenbarger7] were identified as serotype 1c based on their agglutination with MASF 1c. MLST profiles and PFGE patterns indicated that a further 10 rough isolates were also derived from serotype 1c strains. Previously, serotype 1c has been isolated in Bangladesh and Egypt, where it accounted for 8% and 17·5%, respectively, of S. flexneri strains [Reference Ahmed15–Reference Talukder17]. More recently, von Seidlein et al. [Reference von Seidlein18] also reported the isolation of serotype 1c in Indonesia and Pakistan (accounting for 12% and 2% of S. flexneri isolates, respectively). We found that serotype 1c accounted for more than 40% of all S. flexneri strains isolated in Son Tay Province, Vietnam from 1998–1999, making Vietnam the fifth country in which serotype 1c has been identified. This is the first study in which serotype 1c has been identified as the most prevalent S. flexneri serotype in a region. Interestingly, von Seidlein et al. [Reference von Seidlein18] did not identify serotype 1c amongst the 242 isolates they collected between 2000 and 2004 from patients in the central Vietnamese city of Nha Trang. While we identified a single serotype 1c isolate from Nha Trang, serotype 1c appears to be circulating at a very low frequency in this part of the country. There is clearly great variation in the prevalence of different S. flexneri serotypes between different regions of Vietnam.
The level of morbidity caused by serotype 1c strains compared to other S. flexneri serotypes has not been described in the literature. We could not detect any significant difference in the occurrence of clinical symptoms in patients infected with serotype 1c strains compared to patients infected with other S. flexneri serotypes. There was also no significant difference in the occurrence of clinical symptoms in the 10 patients from whom the rough st7-50 isolates were isolated. This may indicate that the loss of O-antigen occurred during the isolation process.
The plasmids observed in serotype 1c isolates were similar to those reported previously for serotype 1 strains isolated in Bangladesh [Reference Talukder13]. The 1·6 MDa plasmid that was present in 72% of serotype 1c isolates from Bangladesh was absent from the Son Tay Province isolates. Two novel plasmids (2·7 MDa and 2·9 MDa) were observed in one of our serotype 1c isolates. These results indicate that the plasmid pool of serotype 1c isolates is largely unchanged across large geographical distances and amongst isolates that are not closely related.
The presence of a large number of different serotype 1c clones, including strains that were ‘unrelated’ to each other, indicates that serotype 1c is well-established in Son Tay Province and that the high proportion of serotype 1c strains identified in this study is a true reflection of this serotype's importance in this region. This is supported by our ongoing isolation and identification of serotype 1c strains from the same region in the years following 1999 (R. M. Stagg, P. D. Cam & N. K. Verma, unpublished data). The identification of a single serotype 1c clone (pattern X5) in two very distant locations (Son Tay Province and Nha Trang), indicates that S. flexneri is transmissible across large distances in Vietnam and that serotype 1c may be an important serotype elsewhere in the country. PFGE also revealed that serotype 1c strains isolated in Vietnam were distinct from serotype 1c strains isolated in Bangladesh, although MLST indicate that they are originally derived from the same parental strain.
The recent increased detection of serotype 1c, both here and elsewhere, is probably due to several factors. First, it is possible that different strategies used to collect isolates favour the isolation of particular serotypes or strains. Epidemiological studies of enteric pathogens are typically retrospective analyses of isolates collected from patients in clinics or hospitals. Hence, reports on the prevalence of different serotypes will be biased, favouring strains that cause severe disease warranting treatment rather than less virulent strains. The isolates studied here were collected following surveillance of children in the community, many of whom may have suffered from disease that was less severe than that experienced by people actively seeking treatment. If serotype 1c rarely causes disease warranting clinical treatment then this could lead to under-reporting of morbidity caused by this serotype. However, analysis of our data indicated that the severity of disease caused by serotype 1c was not significantly different from that caused by other serotypes isolated by the same method.
The lack of widely available detection methods for serotype 1c almost certainly contributes to it being widely ignored. There is anecdotal evidence that isolates that are untypable with commercial sera are often discarded by hospitals, biasing results of epidemiological studies based on these collections.
It is possible that the relative prevalence of serotype 1c is genuinely increasing. This hypothesis is supported by studies performed in Bangladesh, where the serotype 1c monoclonal antibody MASF 1c has been used for serotyping for the past two decades. In the mid-1980s serotype 1c accounted for <1% of S. flexneri isolates, but had increased to 8·2% for the period 1997–2000 [Reference Talukder17, Reference Carlin19]. Changes in serotype prevalence may represent random fluctuations of serotypes over time and region or may result from changed environmental conditions (e.g. widespread antibiotic use) that favour some strains over others.
Our findings confirm that serotype 1c must be regarded as an important S. flexneri serotype and that serotype 1c-specific antibodies should be made widely available to aid in its detection. Our results support the current thinking that vaccines must target a wide range of S. flexneri serotypes if they are to effectively reduce the incidence of shigellosis. Specifically, our data suggests that vaccines designed to reduce the incidence of dysentery in northern Vietnam will have minimal impact unless they target S. flexneri serotype 1c. The emergence of serotype 1c from a seemingly rare S. flexneri serotype to a common and widely distributed serotype, highlights the importance of having tools available for the detection of all bacterial pathogens. Our findings emphasize the value of typing strains with a variety of methods, including using biochemical and genetic typing methods to supplement antigenic typing.
ACKNOWLEDGEMENTS
Thanks are due to Nils Carlin for donating MASF B, MASF 1c and serotype 1c reference strains; Nguyen Thi Gam and Nguyen Thi Be for collecting isolates and clinical data from Son Tay Province; Kaiser Talukder for supplying serotype 1c reference strains, and Gwen Allison for assistance with PFGE.
DECLARATION OF INTEREST
None.









