Article contents
Structural Analysis of Gliding Motility of a Bacteroidetes Bacterium by Correlative Light and Scanning Electron Microscopy (CLSEM)
Published online by Cambridge University Press: 02 February 2022
Abstract
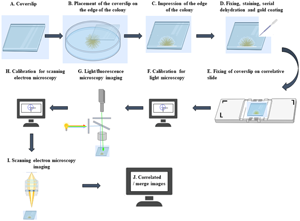
The members of the Bacteroidetes phylum move on surfaces by gliding motility in the absence of external motility appendages, leading to the formation of spreading colonies. Here, the structural features of the spreading colony were assessed in a uranium-tolerant Bacteroidetes bacterium, Chryseobacterium sp. strain PMSZPI, by using correlative light and scanning electron microscopy (CLSEM). We developed a simple and convenient workflow for CLSEM using a shuttle and find software module and a correlative sample holding slide designed to transport samples between the light/fluorescence microscope (LM/FM) and the scanning electron microscope (SEM) to image spreading colony edges. The datasets from the CLSEM studies allowed convenient examination of the colonial organization by LM/FM followed by ultrastructural analysis by SEM. The regions of interest (ROIs) of the spreading colony edges that were observed in LM/FM in the absence and presence of uranium could be re-identified in the SEM quickly without prolonged searching. Perfect correlation between LM and SEM could be achieved with minimum preparation steps. Subsequently, imaging of the correlated regions was done at higher resolution in SEM to obtain more comprehensive information. We further showed the association of uranium with the gliding PMSZPI cells by energy-dispersive X-ray spectroscopy (EDS) attached to SEM.
- Type
- Biological Applications
- Information
- Copyright
- Copyright © The Author(s), 2022. Published by Cambridge University Press on behalf of the Microscopy Society of America
Footnotes
The first two authors contributed equally to this work.
References
- 1
- Cited by





