Health technology assessment (HTA) has been recently defined as the “multidisciplinary process that uses explicit methods to determine the value of a health technology,” aiming at informing decision making “in order to promote an equitable, efficient, and high-quality health system” (Reference O’Rourke, Oortwijn and Schuller1).
In the last two decades, after the conclusion of the Human Genome Project and thanks to the increasing interest in the genomic field, the prospect of integrating genetic determinants for both Personalized Medicine and Personalized Public Health has opened up (Reference Cleeren, Van der Heyden, Brand and Van Oyen2). Genomic medicine represents a key element of personalized medicine, which is informed by individuals’ clinical, genetic, genomic, and environmental information (Reference Ginsburg and Willard3). Moreover, omics technologies are precious tools for personalized decision making (Reference Hoxhaj, Govaerts and Simoens4). Among the multiomics approaches, next-generation sequencing (NGS) is capable of sequencing a predefined set of genes (i.e., panel sequencing), the coding part of genes (i.e., whole exome sequencing, WES) or the whole genome (i.e., whole genome sequencing, WGS). Particularly, WGS is emerging into European healthcare systems albeit still mainly in research setting rather than in clinical practice (Reference Marshall, Bick and Belmont5). The emergence of WGS-based diagnostic tests, as a new opportunity for health care, poses major challenges for the translation of knowledge and technologies from basic science into point-of-care applications (Reference Becla, Lunshof and Gurwitz6). Particular challenges to HTA processes include the potential predictive nature of the risk of disease, the familial implications, laboratory structures (7), as well as the technical and analytical issues specific to WGS (e.g., accuracy and depth of sequencing) and the current difficulties related to the interpretation of the information contained in the genome (Reference Petersen, Fredrich, Hoeppner, Ellinghaus and Franke8).
Nevertheless, the application of diagnostics-based personalized medicine could have the potential to be beneficial to a large variety of stakeholders such as national bodies, healthcare providers, and especially patients. Health systems are therefore pressed from both patients’ and clinicians’ perspectives to adopt these costly technologies (Reference Agyeman and Ofori-Asenso9). Even though WGS could be the promise to shorten the “diagnostic odyssey” experienced by patients affected by rare genetic disorders (10–Reference Wu, McMahon and Lu12), available evidence in the scientific community is still scarce (Reference Bauer, Kandaswamy and Weiss13). A comprehensive assessment is hence needed to provide healthcare commissioners with information on whether introducing WGS adds value into the whole system and it is worth funding (Reference Payne, Eden, Davison and Bakker14). Given the sharp growth in volume and a concurrent plummet in the price of genetic diagnostic tests in recent years, the potential economic assessment is seen as a strategic priority for a debate across HTA organizations worldwide (Reference Becla, Lunshof and Gurwitz6;Reference Husereau, Marshall, Levy, Peacock and Hoch15). HTA reports represent the best tool to support decision makers, by summarizing all the evidence with respect to all the dimensions characterizing the HTA processes.
In light of the novelty of this diagnostic test and the initial interest by national or regional HTA organizations around the globe, the scoping review methodology is suitable to examine emerging evidence when it is unclear whether other more specific research questions can be raised and consequently addressed by a more systematic approach (Reference Armstrong, Hall, Doyle and Waters16).
The aim of this scoping review is to map the available evidence about the use of HTA in the assessment of WGS.
Materials and methods
Study Design and Search Strategy
A scoping review of the literature was conducted to map the assessment of WGS in an HTA perspective, adopting the methodological framework suggested by Arksey and O’Malley (Reference Arksey and O’Malley17) and refinements by the Joanna Briggs Institute (18;Reference Peters, Godfrey and Khalil19).
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) was used to elaborate a comprehensive search strategy necessary to query relevant electronic databases as MEDLINE, Scopus, and EconLit as well as the HTA agencies affiliated with EUnetHTA, HTAsiaLink, and RedESTA. The search was also expanded to EMBASE, the National Health Service’s Economic Evaluation Database (NHS EED), the Center for Reviews and Dissemination (CRD) Health Technology Assessment Database, the Health Technology Assessment bodies listed in the directory of the International Society for Pharmacoeconomics and Outcomes Research (ISPOR), the Health Technology Assessment Central resource center by ISPOR, and to public and/or private insurers as well as by health providers. The full search strategy is reported in the Supplementary Material.
Study Selection
The population, concept, and context (PCC) framework was adopted to establish the eligibility criteria. In particular, inclusion criteria were defined as reports involving a population with suspected genetic disease either in intensive care unit or outpatient settings, describing the use of WGS in the context of HTA organizations or agencies or other bodies worldwide. Studies focusing on patients with any type of neoplasm were excluded given the choice to put the attention on the use of WGS for diagnostic purposes and not for therapeutic ones. The inclusion of potential reports was also restricted according to language (i.e., English), availability of full-texts, and type of articles. Table 1 reports the specific inclusion and exclusion criteria.
Table 1. Inclusion and Exclusion Criteria

PCC, population, concept, and context.
The initial screening by titles and abstracts as well as the final round of screening by full-texts was performed by two independent researchers to determine final eligibility. Any potential disagreements were solved by a third author.
Data Extraction
A data charting form was drafted encompassing report characteristics (i.e., authors’ name, country, year of publication, and title), HTA organization, HTA domains, type of intervention (also including the tests that served as comparator to WGS), population, and setting. The data extraction was conducted by two independent authors.
HTA Classification System and Methodological Characteristics
According to the criteria of the classification system developed by Merlin et al. (Reference Merlin, Tamblyn and Ellery20), the HTA reports were classified into four categories, known as full HTA, mini HTA, rapid review and other. A HTA report is considered full if it met at least seven out of eight criteria, a mini-HTA report had to comply with criteria 1, 2, 4, 6, and 7 while a rapid review had to meet at least criteria 1 and 2. Other reports that did not fit into any of the previous categories were classified as “other.”
Data Synthesis
A preliminary descriptive analysis was performed employing tables and figures to provide summaries regarding the characteristics of each report. Furthermore, a narrative synthesis was conducted taking into account the HTA organizations and the investigated HTA domains.
Results
Reports Selection and Characteristics
Overall, of the 1,794 records retrieved after data deduplication, 1,787 were screened out since they were either studies not including WGS as intervention, studies focusing on different diseases (i.e., neoplasms), or alternatively no structured HTA reports. As a result, seven records (21–27) were included in the review. The full selection process is depicted in Figure 1.

Figure 1. PRISMA flow diagram.
Four HTA reports, drafted over 10 years, were elaborated between 2019 and 2020. Health Quality Ontario (HQO), a Canadian HTA organization, produced the only full HTA report included (21) whereas the Canadian Agency for Drugs and Technologies in Health (CADTH) elaborated two rapid reviews (Reference Young and Argáez24;Reference Herington and McCormack25). The other two were produced by the British National Institute for Health Research (NIHR) and the Belgian Health Care Knowledge Centre (KCE), respectively (Reference Hanquet, Vinck and Thiry22;Reference Beale, Sanderson, Sanniti, Dundar and Boland26). Furthermore, the “other” reports were drafted by the CADTH and the Swedish Agency for Health Technology Assessment and Assessment of Social Services (SBU) in the extent of one report each (23;27).
The scoping review identified that five HTA organizations from four countries elaborated the reports. The seven reports were mainly focused on the evaluation of the clinical utility and cost-effectiveness of genome-wide sequencing as well as informing policy questions by providing analyses of organizational and ethical considerations. Particularly, the report by HQO, focusing on the effectiveness and cost-effectiveness of WGS and people perspectives and experiences about WGS, found that compared with standard genetic testing (e.g., chromosomal microarray, gene panels, or targeted single-gene), WGS has a higher diagnostic yield. Nonetheless, considering also the costs related to the diagnostic tests, using WES as a first- or second-tier test is more cost-effective with respect to standard genetic testing and WGS (i.e., ICER $3,261 per patient). Moreover, the budget impact analysis (BIA) showed that its impact ranged from $4 to $5 million across 5 years. The authors also showed that individuals have consistent motivations for and expectations of obtaining a definitive diagnosis through genome-wide sequencing (21). The first report by CADTH, focusing only on the ethical aspects, found that WGS as well as WES is subject to many of the same ethical concerns as other genetic tests including considerations of respecting individual autonomy through consent processes, balancing harms and benefits with attention to stigmatization, protecting confidentiality while considering how to protect individuals, and how to manage incidental findings. Moreover, it generates novel ethical concerns due to a high volume of incidental findings and the substantial uncertainty inherent to the technology (23). The second report by CADTH, examining the clinical utility, the cost and economic evidence, and the organizational issues of WGS, highlighted that WGS and WES with a rapid turnaround time, compared to standard genetic testing, significantly lower the time to diagnosis and increase the rates of change in medical management. The authors also stated that no evidence regarding the cost-effectiveness and organizational issues of providing rapid turnaround WGS and WES was retrieved (Reference Young and Argáez24). The last report by CADTH, investigating the ethical and social aspects, found that genetic testing is seen by families and clinicians as a valuable tool for determining causal associations for unexplained conditions and to reduce the lengthy diagnostic odysseys (Reference Herington and McCormack25). Furthermore, the report by the NIHR was mainly focused on the current use, cost and economic evidence, and organizational aspects of WGS, WES and targeted gene panels. In relation to the organizational aspects, the main resource-related issues pertaining to service provision are the need for additional computing capacity, more bioinformaticians, more genetic counselors and also genetics-related training for the public and a wide range of staff. The main issues relating to systems and safeguards are giving informed consent, sharing unanticipated findings, developing ethical and other frameworks, equity of access, data protection, data storage, and data sharing. The authors found little published evidence on cost-effectiveness highlighting that the major hurdles to assess cost-effectiveness are the uncertainty around diagnostic yield, the heterogeneity of diagnostic pathways and the lack of information on the impact of a diagnosis on health care, social care, educational support needs and the extended family (Reference Beale, Sanderson, Sanniti, Dundar and Boland26). Besides, the report by the KCE, assessing the same domains of the report by the NIHR, provided a detailed overview of the use of WGS in national (i.e., Belgium) and international clinical practice and of the organization of genetic testing in Belgium. The authors also discussed some organizational issues identifying nine pillars needed to address the challenges related to the introduction of a new technology: infrastructure for WGS; bioinformatics and medical interpretation; data storage; common database of variants; WGS indications and prescription; incidental findings and genetic counseling; financing options; plan for human resources.
The authors put also their attention on the cost and economic evidence reviewing four cost studies: a US study estimated the costs of WGS at $17,620 per test in 2014; Tsiplova et al. running a microcosting analysis, in 2016, estimated a cost of $2,851 (95 percent CI: 2,750, 2,956) and $5,519 (95 percent CI: 5,244, 5,785) for the HiSeq X and HiSeq 2500 platform, respectively; Van Nimwegen et al. in 2016, considering only the direct medical costs, quantified a cost of €1,669 per sample. The last study, conducted in Germany by Plöthner et al. in 2017, was a cost analysis based on direct medical and labor costs. The cost per WGS test was €1,411 with the HiSeq X and €3,858 with the HiSeq 2500 (Reference Hanquet, Vinck and Thiry22). The last report was elaborated by the SBU and it considered the social and ethical aspects of WGS and WES compared to karyotyping. The authors found that the adoption of NGS could introduce ethical problems regarding how to sufficiently explain the method and the expected results, how to determine who should be analyzed and what kind of information should be provided to whom (27). Additional characteristics of all included reports are provided in Table 2.
Table 2. Summary Characteristics of Included Reports

aCGH, array comparative genomic hybridization; CADTH, Canadian Agency for Drugs and Technologies in Health; CMA, chromosomal microarray; HQO, Health Quality Ontario; HTA, health technology assessment; ICU, intensive care unit; KCE, Belgium Health Care Knowledge Centre; NICU, Neonatal Intensive Care Unit; NIHR, National Institute for Health Research; N.S., not specified; PICU, Pediatric Intensive Care Unit; QF-PCR, quantitative fluorescence-polymerase chain reaction; SBU, Swedish Agency for health technology assessment and assessment of social services; TGP, targeted gene panel; WES, whole exome sequencing; WGS, whole genome sequencing.
HTA Classification System and Methodological Characteristics
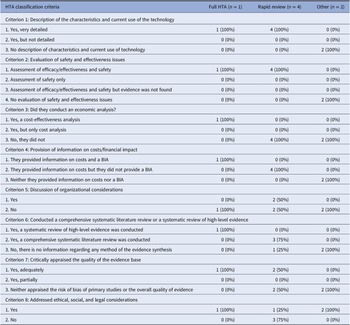
Table 3 shows the results of the classification of HTA reports based on the classification system devised by Merlin et al. (Reference Merlin, Tamblyn and Ellery20).
Table 3. Results of HTA Classification

BIA, budget impact analysis; HTA, health technology assessment classification criteria to define the type of HTA reports were retrieved from Merlin et al. (Reference Merlin, Tamblyn and Ellery20).
The only full HTA report met seven out of eight criteria, including a detailed discussion about the costs, cost-effectiveness, budget impact and also ethical and social aspects, which is optional. On the contrary, the report did not take into account organizational considerations. The report systematically mapped the scientific literature and assessed the methodological quality of primary studies. The four rapid reviews performed a review of the literature but did not appraise the risk of bias or the overall quality of evidence. All provided information on costs. Moreover, one of the rapid reviews investigated social aspects. The reports classified as “other” evaluated only ethical considerations related to the adoption of WGS.
Discussion
This study was intended to map the current evidence about the adoption of WGS in the diagnosis of genetic diseases drafted by the HTA agencies worldwide.
A still low number of HTA assessments of WGS was found in line with the novelty of the investigated topic (Reference Kumar, Cowley and Davis28;Reference Schwarze, Buchanan, Taylor and Wordsworth29). In addition, few countries are elaborating HTA reports for WGS. The Canadian HTA agency drafted approximately half of the included reports. None of the reports were produced in lower-middle- and low-income countries. Typically, the public sector of these emerging markets have low access to innovative medical technologies, such as WGS, due to an inadequate number of equipped laboratories with the capacity to screen several individuals, the lack of health management information systems and, altogether, limited resources allocated to the public health system (Reference Tekola-Ayele and Rotimi30;Reference Hetu, Koutouki and Joly31).
The results of the study show a very limited amount of robust and complete HTA reports for WGS, largely due to the lack of systematic literature review, scarcity of methodological quality appraisal of retrieved evidence, and absence of considerations about economic issues such as BIA and cost-effectiveness models. The main reason could be the current difficulties in defining the main assumptions of the economic evaluations and also in the retrieval of necessary cost and effectiveness data. Considering the latest available evidence (Reference Li, Vandersluis and Holubowich32;Reference Wu, Balasubramaniam and Rius33), few full economic evaluations were conducted to investigate the cost-effectiveness of WGS with respect to WES and standard testing (and other scenarios as the comparison between WGS and second-tier WES) sharing quite similar methodological assumptions as the report by the HQO. The two cost-effectiveness analyses were based on few main assumptions as the population considered (i.e., only patients with suspected genetic disorders) and the adoption or exclusion of specific parameters in the analysis (e.g., currently it is difficult to detect Fragile X syndrome with genome-wide sequencing so neither the cost of such testing nor the related diagnostic yield was included and also the costs of returning secondary findings were excluded due to the inherent challenges of modeling the benefits and costs of unpredictable secondary findings). Both the evaluations compared WGS with respect to WES and standard genetic testing assessing the diagnostic yield, the number of changes in medical management and the turnaround time as main outcomes and the cost of genetic (e.g., WGS, WES, and standard testing cost) and nongenetic diagnostic tests (e.g., biochemical/metabolic workup, neuroimaging, and laboratories tests), pretest and posttest genetic services (e.g., genetic counseling), equipment costs, personnel costs, and supplies costs as the main cost items.
Only the report by the HQO performed a full economic evaluation sharing similar assumptions and considering the same outcome measures of the studies cited above. However, its findings differ from those of the two studies possibly because of the setting and the time-period in which the assessment was conducted. Another reason related to the lack of robust and complete HTA reports may be due to the fact that, as figured out by INAHTA members in the top ten challenges to produce an HTA report, an increasing demand for HTAs was frequently followed by a request for larger speed leading to the production of more rapid HTAs (Reference O’Rourke, Werkö, Merlin, Huang and Schuller34). In addition, INAHTA members stated that adopting evidence grading system in the formulation of recommendations for policy makers was a hurdle. This is consistent with the findings of the present review, since almost all the included reports did not appraise the methodological quality of primary studies or the quality of evidence with the proper frameworks as the Grading of Recommendations Assessment, Development and Evaluation (GRADE) and the Cochrane risk-of-bias scale tools.
Moreover, although health economic evaluations and BIA are highly recommended and suggested by the literature (Reference Kristensen, Husereau and Huić35;Reference Stephens and Hanke36) to enrich the multidisciplinary assessment, they were not a common practice in the HTA reports of WGS identified in this review.
Organizational, social, and ethical issues such as the minimum genetic laboratory requirements, the available genetic tests, the required essential personnel, the training of personnel, the disclosure of incidental and/or secondary findings, patient privacy and risk of discrimination, were addressed in the full HTA report but not in most of the rapid reviews and other reports. According to the literature, the HTA process, in its assessments, does not still deepen context-dependent aspects (e.g., organizational, social, and ethical factors) that may reflect a linkage between HTA and health policy practice (Reference Stephens and Hanke36–Reference Pfadenhauer, Rohwer and Burns39).
In light of the above, the failure to include these context-dependent aspects in the multidisciplinary analysis may result in policy makers relying solely on clinical evidence preventing HTA evaluations from bridging this knowledge-action gap (Reference Velasco Garrido, Gerhardus, Røttingen and Busse37).
Specifically, policy makers may fail in the application of HTA reports of WGS as they might not take into account the context in which it would be adopted and executed in healthcare practice (Reference Sharma, Choudhury and Kaur40).
The joint reading of the review findings leads to a main suggestion. It is of paramount importance to stimulate a critical reflection in the way of producing, implementing and executing HTA reports with respect to the three levels characterizing the decision-making process in the healthcare system. At the first level (i.e., macro level), researchers and scholars, in national or regional HTA organizations, should deepen all the aspects related to the WGS and not only those regarding the clinical evidence adopting principles of accuracy, comprehensiveness and transparency in drafting HTA reports as well as assess the sustainability of this technology. At the second level (i.e., meso level), healthcare providers should manage the procurement and surveillance of WGS use. At the third level (i.e., micro level), health professionals (e.g., geneticists) should pay attention to the information included in the HTA reports of WGS. As a result, HTA reports will be increasingly accurate and complete in the areas of investigation, and as such, will better be able to answer emerging questions and allow evidence-based decisions to be made.
The findings of this study and the preliminary conclusions must be considered in light of the study’s weaknesses. The main limitation was the limited number of retrieved reports and the source of evidence, mainly based on the grey literature. However, the accurate methodology, widely applied in the literature, is the strength of this study. Another caveat concerned the restriction of the search strategy to English, hence potentially leading to language bias and exclusion of eligible reports issued in other languages.
Furthermore, despite the wide availability of checklists and guidelines about the design, interpretation, and reporting of several types of reviews (Reference Page, McKenzie and Bossuyt41;Reference Page, Moher and Bossuyt42), there is still scarcity of structured guidance for scoping reviews (Reference Arksey and O’Malley17;Reference Tricco, Lillie and Zarin43), particularly related to the quality assessment process. Scoping reviews do not formally include the evaluation of the methodological quality of the included studies and, consequently, scholars might not be conscious of the potential biases that may stem, resulting in highly questionable findings.
Further studies are needed to also consider the other domains of HTA (e.g., cost-effectiveness, organizational, social, and ethical) given their importance in the formulation of health policies. Additional research, from an HTA perspective, is also required to fully prove the sustainability of WGS in health systems and, thus, to pave the way for the establishment of reimbursement for its provided services.
Conclusions
According to current information, few HTA organizations are drafting reports for WGS in the diagnosis of genetic diseases, focusing mainly on cost-effectiveness, organizational and ethical issues. Lessons learned from this study reinforce the need to encourage a critical reflection during the elaboration of HTA reports assessing the use of WGS in the diagnosis of rare genetic diseases in order to encompass all relevant domains necessary for the multidisciplinary assessment characterizing the HTA process. This allows to accurately steer choices of decision makers in the establishment of priorities for research and policy, in case of widespread adoption, and reimbursement rates of this innovative health technology.
Supplementary Material
The supplementary material for this article can be found at https://doi.org/10.1017/S0266462322000496.
Funding Statement
This research received no specific grant from any funding agency, commercial, or not-for-profit sectors.
Conflicts of Interest
The authors declare that they have no conflict of interest.