INTRODUCTION
Since the 1990s, the epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) infections has changed markedly owing to the emergence of community-associated MRSA (CA-MRSA) infections as a serious health problem in many parts of the world [Reference Yamamoto1]. Initially CA-MRSA strains were identified as being responsible for the increase of staphylococcal infections in communities worldwide, but now they have expanded to also become a causative agent of nosocomial infections and thus the distinction between hospital- and community-acquired strains is becoming less clear [Reference Yamamoto1]. Methicillin resistance in MRSA and coagulase-negative staphyloccocci (CoNS) is encoded by the mecA gene which is located within the staphylococcal cassette element (SCCmec) integrated in the bacterial chromosome [Reference Ito2–Reference Barbier4]. Genotypic and phenotypic differences between CA-MRSA and hospital-acquired MRSA (HA-MRSA) strains are well recognized and include the type of SCCmec element, their virulence factor profile, and wider antimicrobial susceptibility [Reference Deurenberg and Stobberingh5].
Nasal carriage of MRSA by hospital staff in Jordan was first reported in the early 1990s [Reference Na'was and Fakhoury6]. Subsequently, high rates of S. aureus (40%) and MRSA (19%) were recorded in young Jordanian adults in the community [Reference Al-Zu'bi, Bdour and Shehabi7], which were somewhat higher than the rate (22·7%) found in a survey of 132 healthy students published 4 years earlier [Reference Daghistani, Issa and Shehabi8]. Recently, Khalil et al. [Reference Khalil9] reported ST80-MRSA-IV as the dominant clonal type in hospitalized children in Jordan, although the extent of MRSA and MR-CoNS carriage in the healthy Jordanian community and the molecular characterization of these strains remain inadequately investigated.
A major risk factor for infections with methicillin-resistant staphylococci (MRS) is the carriage of these microorganisms at different body sites. Indeed their co-existence might facilitate the exchange of mobile resistance and other genetic elements and there is some evidence to suggest that MR-CoNS may act as a source of SCCmec for MRSA [Reference Barbier4, Reference Diep10, Reference Archer and Niemeyer11]. Although MR-CoNS colonization might be correlated with the emergence of MRSA, there are few epidemiological studies examining co-colonization in the healthy population for these two groups of organisms and it has been shown that there is no significant difference in terms of nasal carriage for S. aureus and MRSA [Reference Chen, Chen and Huang12] in pre-clinical and clinical university students; therefore university students are a suitable representative group on which to draw conclusions on the spread of MRSA in the wider community population [Reference Chen, Chen and Huang12, Reference Prates13].
In the current study we aimed to determine the skin and nasal carriage rates of MR-CoNS and MRSA in healthy preclinical students and faculty staff in the Jordanian community. A secondary aim was to obtain baseline epidemiological data to inform the design and implementation of appropriate infection surveillance and control practices. Third, we investigated all isolates from the survey in detail with regard to their molecular genetic types and virulence gene profiles, and wider antimicrobial susceptibility.
METHODS
Subjects and data collection
During June 2009 to December 2009, a total of 454 microbiological samples were collected from 227 apparently healthy volunteer pharmacy students and employees at the Faculty of Pharmacy, The University of Jordan. Most (89%) of the students had not been exposed to clinical training within the 2 months prior to sampling. Written consent was obtained prior to commencing the study which complied with ethical guidelines of experimental approval (number 14/2007–2008) obtained from the Scientific Research Council at the Deanship of Academic Research, The University of Jordan. The following data were collected: demographic characteristics, medical history including previous and recent hospitalization (within a year), antibiotic consumption, and family member being a healthcare worker.
Sampling and bacteriological identification
The dorsum of the forearm (1 cm2) and the anterior nares of each volunteer subject were sampled [Reference von Eiff14] using sterile dry cotton swabs (Deltalab, Spain) pre-moistened with brain heart infusion (BHI) broth (Oxoid, UK) containing 6·5% NaCl. Each swab was immersed in the same broth and incubated at 37 °C for 2 h. The broth was subcultured onto mannitol-salt-agar (Oxoid) supplemented with methicillin (10 mg/l), and incubated at 35 °C for 24–72 h. Each distinctive colony morphotype was selected, Gram-stained and biochemically identified which included catalase (Merck, Germany), tube coagulase (Remel, UK) and DNase tests (Oxoid). Presumptive MRSA isolates (n = 37) and randomly selected MR-CoNS isolates (n = 51) were analysed using MICROBACT™ 12S (Oxoid) according to the manufacturer's recommendations. Isolates were stored in BHI broth supplemented with 10% glycerol, at −70 °C.
DNA extraction and PCR procedures
LB broth (Merck) overnight cultures of the MRSA and MR-CoNS isolates were prepared. Chromosomal DNA was extracted according to the manufacturer's instructions using the Wizard Genomic DNA purification kit (Promega, USA), with lysostaphin (Sigma, USA) at 25 μg/ml for the lysis step. MRSA isolates (n = 37) and MR-CoNS isolates (n = 298) were analysed by PCR in a PTC-100 thermocycler (MJ Research, USA) for the presence of nuc gene [Reference Brakstad, Aasbakk and Maeland15] and tuf gene [Reference Martineau16], respectively. The multiplex PCR method [Reference Milheirico, Oliveira and de Lencastre17] was used for the determination of SCCmec types for all methicillin-resistant isolates and subtyping of SCCmec type IV was performed according to Milheirico et al. [Reference Milheirico, Oliveira and de Lencastre18]. All MRSA isolates were analysed for the presence of Panton–Valentine leukocidin (PVL) encoding genes (lukS-PV, lukF-PV), γ-haemolysin [Reference Lina19], toxic shock syndrome toxin (tst) and enterotoxin (sea-see, seg-sej, sem-seo) genes [Reference Jarraud20].
The following were used as reference strains, MRSA strains: COL, ANS46, MW2, 8/6-3P, Q2314, JCSC4469, HAR22, WIS, HDE288 [Reference Milheirico, Oliveira and de Lencastre17, Reference Milheirico, Oliveira and de Lencastre18], Bk2464 [Reference Oliveira, Tomasz and de Lencastre21].
spa typing of Staphylococcus aureus
The polymorphic X region of the spa gene was amplified according to Shopsin et al. [Reference Shopsin22]. The PCR products were purified using a PCR product purification kit (Qiagen, Germany) and DNA sequenced (Macrogen, Korea). The obtained sequences were analysed using RidomStaphType software version 2.2.1 (Ridom GmbH, Germany) and spa types were assigned according to the spa server database (http://spaserver.ridom.de). The relationship of the spa types was analysed using the BURP (based upon repeat patterns) algorithm with default parameters within the RidomStaphType software package.
Antibiotic susceptibility testing
Antibiotic susceptibility of isolates was determined using a disc diffusion method as described previously [23] with the following antibiotics (Oxoid): penicillin (10 IU), oxacillin (1 μg) cefoxitin (30 μg), vancomycin (30 μg), gentamicin (10 μg), erythromycin (15 μg), clindamycin (2 μg) norfloxacin (10 μg), tetracycline (30 μg) and linezolid (30 μg). MR-CoNS isolates, which were erythromycin resistant and appeared to be susceptible to clindamycin, were further tested for possible inducible clindamycin resistance as described previously [23].
Statistical analysis
Categorical variables between groups were compared by means of Pearson's χ2 test [exact significance (two-sided)] using SPSS version 16 (SPSS Inc., USA) to evaluate the relationship between risk factors with the carriage rate of MRS, MRSA, MR-CoNS and MRSA + MR-CoNS co-colonization. A P value of <0·05 was defined as significant.
RESULTS
Population characteristics
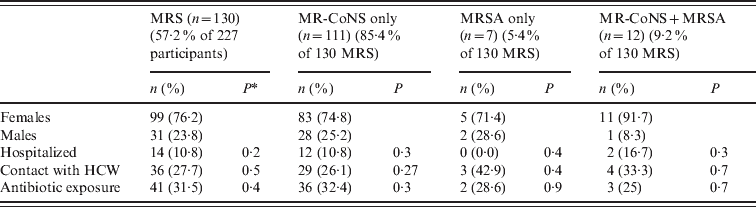
The basic demographics of the volunteers screened by nasal and skin swabs are presented in Table 1. Of 227 study participants, 173 (76·2%) were females (age range 19–26 years) and 54 were males (23%, age range 20–38 years). Twenty participants (8·8%) had been recently hospitalized. The majority (216, 95·1%) were pharmacy students and 11 (4·8%) were faculty members; 67 (29·5%) individuals had a relative working in healthcare and 67 (29·5%) had exposure to antibiotics during the study period.
Table 1. Demographics of 227 healthy volunteer subjectss and distribution of MRS, MR-CoNS only, MRSA only and MR-CoNS + MRSA isolates among these subjects

MRS, Methicillin-resistant staphylococci; MR-CoNS, methicillin-resistant coagulase-negative staphylococci; MRSA, methicillin-resistant Staphylococcus aureus; HCW, healthcare worker.
* Statistically significant trend (*P < 0·05) calculated by Pearson's χ2 test.
Carriage rates of community MRS
Initial biochemical identification of the colonies grown from 454 samples resulted in the isolation of 335 MRS from 130/227 (57·2%) subjects (Table 1). Of these, 37 isolates were MRSA and were recovered from 16 females and three males (Table 2). The prevalence of MRSA nasal carriage was 7·5% (17/227) and 8·4% (19/227) for both nasal and extranasal colonization (Table 2). The remaining isolates (n = 298) were identified as MR-CoNS and were isolated from 123 participants (carriage rate 54·2%), predominantly from nasal samples (Table 2). Of the 12 individuals with MRSA and MR-CoNS carriage, nasal co-colonization was detected in eight. None of the studied risk factors was significantly correlated with carriage in either group of staphylococci (Table 1).
Table 2. Distribution of MRSA and MR-CoNS isolates among 227 study volunteer subjects

MRSA, Methicillin-resistant Staphylococcus aureus; MR-CoNS, methicillin-resistant coagulase-negative staphylococci.
The species-specific tuf gene was identified in 152/298 MR-CoNS isolates from 87 subjects and 51 of the tuf-positive isolates were additionally identified by MICROBACT. Eighty percent were MR-S. epidermidis (MRSE) with the remainder being S. capitis (7·8%), S. chromogenes (3·9%), S. hominis (3·9%), S. cohnii (2%) and S. haemolyticus (2%).
Antimicrobial susceptibility
All MRSA isolates were resistant to all β-lactam antibiotics, but susceptible to all other agents tested. The majority of tuf-positive MR-CoNS isolates were susceptible to gentamicin (97·4%), tetracycline (90·8%), norfloxacin (84·9%), and clindamycin (79·6%). Furthermore, 52·6% showed resistance to erythromycin. Inducible clindamycin resistance was detected in 11 (7·2%) of 54 clindamycin-sensitive/erythromycin-resistant MR-CoNS isolates. All MR-CoNS were susceptible to vancomycin and linezolid.
SCCmec types for MRSA and MR-CoNS
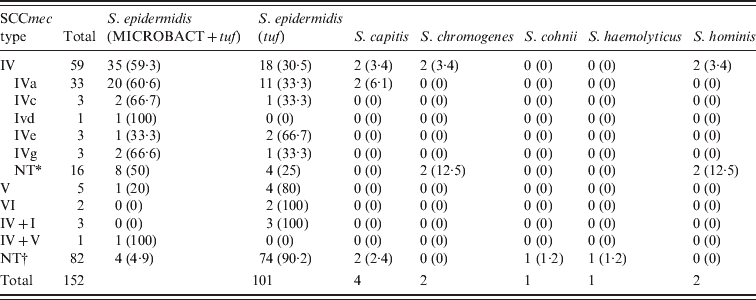
All MRSA isolates harboured the SCCmec type IV element of subtype e (81%) and subtype c. By contrast, MR-CoNS displayed great variability in SCCmec types (Table 3). Sixty-six of 152 isolates were of a single type comprising type IV (n = 59, of which 53 were MRSE), type V (five MRSE) and type VI (two MRSE). Four isolates harboured two types, IV + I (three MRSE), and IV + V (one MRSE). The SCCmec element in 82 of MR-CoNS (78 MRSE) could not be identified by PCR. The identified SCCmec IV subtypes in MR-CoNS were: subtype a (56%), subtype c (5%), subtype d (1·7%), subtype e (5%), subtype g (5%) or untypable (27·1%). For non-S. epidermidis species only MR-CoNS-IV or MR-CoNS-NT were recognized. Significantly, 3/8 subjects colonized with both groups of staphylococci yielded isolates harbouring SCCmec type IV. The other five subjects were co-colonized by MRSA-IV either with MRSE and/or MR-CoNS of SCCmec types V and IV + I.
Table 3. Distribution of SCCmec elements among 152 tuf-positive MR-CoNS from study volunteer subjects

Values given are n (%).
* NT, Untypable by Milheirico et al. [Reference Milheirico, Oliveira and de Lencastre18].
† NT, Untypable by Milheirico et al. [Reference Milheirico, Oliveira and de Lencastre17].
spa types of MRSA and virulence factors
The spa type of the 37 MRSA isolates and their associated virulence factors are presented in Table 4. A single novel spa type, designated t9519, accounted for 76% of the isolates and only two other spa types were identified, i.e. t223 (14·7%) and t044 (5·9%). Two isolates were positive for PVL encoding genes lukS-PV-lukF-PV and belonged to SCCmec IV subtype c, spa type t044. All isolates harboured γ-haemolysin and tst genes, and were enterotoxin-gene positive with the most common being seb (97·3%), seo (94·6%) and sei (91·9%). All isolates were negative for other enterotoxin genes tested: sea, sec, sed, see, seh, and sej. The PVL-positive strain carried only the seb gene.
Table 4. Resistance element, spa type and virulence gene profile patterns of MRSA carriage isolates

* Unable to identify the spa type.
† Untypable by Milheirico et al. [Reference Milheirico, Oliveira and de Lencastre18].
DISCUSSION
Jordan had been reported as being among the countries with hyperendemic antimicrobial resistant bacterial strains. Previously, the Antibiotic Resistance in the southeastern Mediterranean (ARMed) project, reported Jordan as the country with the highest prevalence of significant clinical MRSA infections among the Mediterranean countries [Reference Borg24, Reference Borg25]. This is expected to be correlated to antimicrobial misuse and overuse [Reference Al-Bakri, Bustanji and Yousef26, Reference Al-Momany27] and probably also to a high carriage rate of this microorganism. Surprisingly, we were unable to identify epidemiological factors in this study population, including antibiotic exposure or close contact with a healthcare worker, that were associated significantly with carriage of either MRSA or MR-CoNS. Of interest, the reported nasal carriage rate of MRSA found here remains unchanged from that reported previously (7·6%) in non-medical Jordanian university students [Reference Al-Zu'bi, Bdour and Shehabi7], whereas the geographically adjacent countries of Lebanon and Saudi Arabia have reported markedly lower carriage rates of 1·6% and 1·3%, respectively [Reference Halablab28, Reference Alghaithy29].
To the best of our knowledge, the present study is the first to comprehensively assess the genotypic and phenotypic characteristics of MRSA isolated from healthy Jordanians in the community. Genotypically, they belonged to SCCmec type IV, a type that is often associated with CA-MRSA infections, and is also associated with a minority of hospital strains including some reported from Jordan [Reference Daghistani, Issa and Shehabi8, Reference Khalil9]. This might imply that some strains are circulating and disseminating between the hospital and community.
We report here the identification of a novel spa type (t9519) of MRSA with the methicillin resistance element SCCmec IVe as the predominant type carried by healthy Jordanians. BURP analysis revealed a close relationship to t012 which is a common spa type in Europe (spaserver.ridom.de). Interestingly, t044, which is a widely distributed spa lineage of HA-MRSA in European [Reference Deurenberg and Stobberingh5] and some Middle Eastern countries, including Jordan [Reference Khalil9, Reference Tokajian30], was found in the present study in two healthy individuals.
The susceptibility profiles of the MRSA isolates recovered in the present study are also consistent with a community origin being susceptible to all antimicrobials with the exception of the β-lactams tested [Reference Yamamoto1]. It has been suggested that antimicrobial susceptibility-based classification of CA-MRSA lacks sensitivity and the widely used marker of ciprofloxacin susceptibility may miss approximately one-third of these strains [Reference Otter and French31]. Nevertheless, PVL, a common marker of CA-MRSA, was quite rare in our isolates in accord with reports from both Japan and Ireland [Reference Takizawa32, Reference Rossney33]. The carriage of at least three virulence factor encoding genes suggests that the majority of our MRSA isolates have the potential to cause infections as the combinatory effect of virulence factors is strongly linked to their potential involvement in severe infections [Reference Peacock34].
We present, for the first time, data on the prevalence and molecular epidemiology of MR-CoNS in healthy Jordanians. Consistent with previous reports, the SCCmec elements in MR-CoNS exhibited genetic diversity where some isolates harboured two SCCmec types and others were untypable [Reference Garza-Gonzalez3, Reference Zong, Peng and Lü35]. SCCmec type IV, a relatively common type in MR-CoNS of community origin [Reference Barbier4, Reference Zong, Peng and Lü35], prevailed among our tuf-positive MR-CoNS isolates which proved to be mostly MRSE [Reference Ibrahem36]. In addition other MR species commonly isolated from human clinical specimens, such as S. capitis, S. cohnii, S. haemolyticus and S. hominis, were identified as nasal and skin colonizers. Of note, in our community pool of MR-CoNS isolates, MRSE-IV and MRSE-NT prevailed, and co-harbouring of different SCCmec elements (MRSE-IV + V, MRSE-IV + I) was observed, a finding consistent with MRSE genome plasticity in the community [Reference Lebeaux37]. In agreement with Garza-Gonzalez et al. [Reference Garza-Gonzalez3], antibiotic susceptibility patterns in MR-CoNS isolates are diverse and this is clearly associated with the heterogeneity of SCCmec types present in these strains.
Resistance to β-lactams in MR-CoNS is encoded by SCCmec elements which have high homology with certain types of SCCmec of MRSA [Reference Barbier4]. Such high homology might suggest a common origin and indicate a probable horizontal cross-transmission of resistance genes upon co-colonization [Reference Diep10, Reference Archer and Niemeyer11]. Thus, the sharing of the same SCCmec type between most of the co-colonized MRSA and MR-CoNS, might suggest in vivo horizontal gene transfer but this would need to be confirmed by further genetic characterization not only of these groups but also of co-resistant methicillin-sensitive S. aureus.
This study has some limitations. We were not able to estimate the true prevalence of CA-MRSA infection in the general Jordanian population and wider based surveillance studies in different sections of local and regional geographical sectors are warranted to achieve this. Furthermore, the socio-demographic characteristics and the risk factors associated with MRSA acquisition were not analysed or compared with non-colonized subjects. Due to the spread of various MRSA clones between the community and hospital [Reference Song38], and across national boundaries [Reference Song38], a full molecular characterization of strain lineages by the internationally validated MLST system in order to define them in a global epidemiological context is of importance in both the clinical and infection control setting [Reference Mediavilla39].
As successfully demonstrated in countries or regions with low, decreasing or stabilized MRSA infection rates [Reference Ammerlaan40], it is of utmost importance to develop comprehensive surveillance and prevention strategies that will lead to effective and robust control not only of MRSA but also other multidrug-resistant infections. Major elements of such strategies include implementation of active screening procedures adapted to the specific endemic situation, adherence to basic infection control practices, introduction and control of microorganism-specific hygiene measures, and antibiotic control programmes [Reference Huskins41].
In conclusion, this study is the first to demonstrate a high incidence of MRS nasal and skin carriage in healthy Jordanians, a finding consistent with a community origin for these organisms. We found a high prevalence of MRSA in the community compared to other countries in the region and that most isolates are of the same genotype and share the same β-lactam antimicrobial resistance mechanism. In addition we identified nasal co-colonization with both MRSA and MR-CoNS with highly similar resistance genes in a small number of subjects which raises the possibility of horizontal resistance gene transfer between the different groups of staphylococci.
ACKNOWLEDGEMENTS
We sincerely thank Dr C. Milheiriço (Laboratory of Molecular Genetics, Universidade Nova de Lisboa, Portugal), Dr H. de Lencastre (Laboratory of Microbiology, The Rockefeller University, New York, USA), Dr T. Ito (School of Medicine, Juntendo University, Tokyo, Japan), Dr S. Boyle-Vavra, Dr R. Daum (Department of Pediatrics, Section of Infectious Diseases, University of Chicago, USA) and Dr W. B. Grubb (Faculty of Health Sciences, Curtin University, Australia) for kindly providing the prototype and reference strains used in this study. The project was funded by the Deanship of Academic Research, The University of Jordan (grant no. 14/2007–2008) and the Scientific Research Fund, Ministry of Higher Education (grant no. 3/13/2008). Professor H. Al-Hadithi was on sabbatical leave while supervising and running this study. She is currently working in the Department Biology, Faculty of Science, University of Basrah, Iraq.
DECLARATION OF INTEREST
None.