The silencers: Small RNAs in parasitic nematodes – forms and functions
The latest Paper of the Month for Parasitology is Small RNAs in parasitic nematodes – forms and functions
How do parasitic roundworms, also known as nematodes, develop and adapt to the conditions within the host? And can we exploit this knowledge to develop new ways to control parasitic infections?
To develop, survive and reproduce, parasites need to switch genes on and off in a co-ordinated manner. The mechanisms for regulating gene expression are complex but our understanding of this has greatly progressed thanks to analysis of genome information. Surprisingly, in many nematodes there are a similar number of genes coding for proteins as found in humans (approximately 20,000) and in both, many regions of their genomes act as regulators of these genes, coordinating when, where, and how much of them is expressed.
As organisms have become more complex, gene expression requires more sophisticated regulation: broadly speaking, it can be divided into three distinct mechanisms. First, specific enzymes carry out a process called ‘chromatin remodeling’ where they alter the structure of DNA and ‘open it up’ to allow gene expression. Second, proteins known as transcription factors are involved in transcribing this ‘open’ DNA into messenger RNA (mRNA). A third mechanism of gene regulation, which is the focus of this review, governs the stability of the mRNA molecule after it is made. This is known as post-transcriptional regulation, and involves the recognition of mRNA sequence(s) by regulatory RNA molecules. These regulatory RNAs bind to their target mRNA, together with proteins called Argonautes. As a result, the mRNA is degraded, and the gene is ‘silenced’. While the outcome is the same, post-transcriptional regulation can occur via slightly different mechanisms employing different regulatory RNAs. We review current information on these different small RNA types in parasitic nematodes: microRNA (miRNA), Piwi-interacting RNA (piRNA) and small interfering RNA (siRNA) and outline how they function.
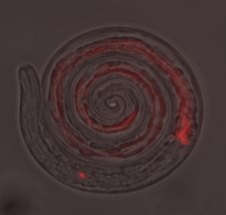
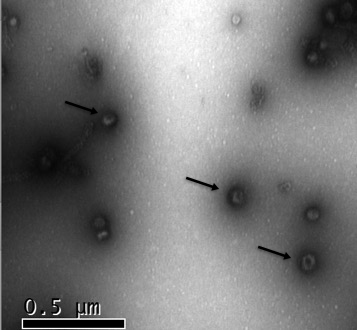
Figures: A) Ingestion of red fluorescent siRNA into the intestine of a larval stage of the parasitic nematode Haemonchus contortus. B) Arrows point to small extracellular vesicles (EV) released by adult stage H. contortus maintained in vitro.
Small RNAs were first identified in the free-living nematode Caenorhabditis elegans, and by comparing genome sequences, it’s been possible to identify small RNAs and the machinery needed for their function in at least some parasitic nematodes. Their conservation indicates fundamental roles, such as miRNAs regulating development through different lifecycle stages. Sequence data also indicate the evolution of novel parasite miRNA sequences, particularly in adult worms, suggesting roles in reproduction and/or in modulating host gene expression. The exciting discovery that small RNAs are packaged into tiny vesicles released by parasites that can be taken up by host cells has opened up new ideas on how parasites may manipulate host immune genes to benefit their survival. piRNAs and siRNAs are crucial in protecting the nematode genome from deleterious sequences, including mobile genetic elements and RNA viruses.
C. elegans small RNA research has been ground-breaking in revealing the important roles that miRNAs and siRNAs play in regulating gene expression in complex organisms, from nematodes to humans. We end the review by discussing further studies to advance our understanding of small RNAs. This exciting avenue of research has the potential to identify new methods to interfere with parasite survival, and thus new ways to control parasitic infections.
The article “Small RNAs in parasitic nematodes – forms and functions” by Collette Britton, Roz Laing and Eileen Devaney, published in Parasitology, is available free for a month.






