Malacosporean myxozoans exploit a diversity of fish hosts
The latest Paper of the Month for Parasitology is ‘Malacosporean myxozoans exploit a diversity of fish hosts’
by Juliana Naldoni, Edson A. Adriano, Ashlie Hartigan and Beth Okamura
Parasitic diseases can represent major challenges for conservation and wildlife management on the one hand, and food production and recreational activities on the other. Critical to mitigating parasitic diseases is knowledge of parasite diversities and the hosts they exploit. As in mammals and birds, fish from natural and farmed environments harbour many parasitic infections, including various protozoans, platyhelminths (flukes and tapeworms), nematodes (roundworms), annelids (leeches), and even cnidarians! Free-living cnidarians are familiar animals, such as jellyfish and sea anemones, but their parasitic relatives comprising the Myxozoa are poorly known. Myxozoans are tiny, morphologically-simplified cnidarians with complex parasitic life cycles, occurring as internal parasites of invertebrate and vertebrate hosts (especially fish). There are two groups of myxozoans – the Myxosporea and the Malacosporea. Myxosporeans use annelids as invertebrate hosts and mainly fish as vertebrate hosts. Around 2500 myxosporean species have been described and some are important fish pathogens, for example causing whirling disease, enteromyxosis, and enteronecrosis. Malacosporeans use freshwater bryozoans as invertebrate and fish as vertebrate hosts. Only a handful of malacosporean species have so far been formally described while another 20 have been detected by their DNA sequences. The most well-known species is Tetracapsuloides bryosalmonae, the causative agent of the devastating and emerging proliferative kidney disease (PKD) of wild and farmed salmonid fishes.
The aim of this study was to investigate malacosporean fish host diversity by sampling for infections in a range of freshwater fish species from geographically distinct sites. Malacosporean infections were detected by DNA sequencing and examined using light and transmission electronic microscopy (TEM). Our DNA analysis revealed five malacosporean species infecting 83 of 256 fish belonging to nine different families. A range of fish from the UK and Switzerland harboured infections (brown trout, rainbow trout, white fish, dace, roach, gudgeon and stone loach). Critically, TEM analysis revealed parasite developmental stages in kidney tubules of four species (brown trout, white fish, roach and stone loach), providing evidence that these fish are indeed true hosts. Our results expand the inferred range of malacosporean fish hosts in Europe and confirm that malacosporeans are capable of exploiting not just fish in the family Salmonidae but also fish in at least two other families – the Cyprinidae and Nemacheilidae.
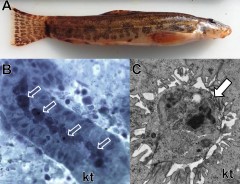
Figure 1. Buddenbrockia sp. 2 discovered infecting kidney of stone loach. A – Stone loach (Nemacheilidae) collected in the River Stour, UK. B – Light microscopy showing the parasite (arrows) in the fish kidney tubule (kt). Scale bar: 10 µm. C – TEM showing development of an infectious spore (arrow) in the fish kidney tubule (kt) confirming stone loach as a true host. Scale bar: 1 µm.

Juliana Naldoni and Edson A. Adriano are in the Department of Ecology and Evolutionary Biology at the University of São Paulo-UNIFESP, Brazil. Ashlie Hartigan is based in the Faculty of Life Sciences & Medicine, King’s College London and the Department of Life Science at the Natural History Museum, London, where Beth Okamura is also based. This research was funded by São Paulo Research Foundation-FAPESP, grants #2016/22047-7 and #2016/08831-7 and was conducted at the Natural History Museum, London.
The paper ‘Malacosporean myxozoans exploit a diversity of fish hosts’ is available free for a month.






