Hospital-acquired pneumonia (HAP) is the most common hospital-acquired infection, representing ∼22% of all hospital-acquired infections. Reference Magill, O’Leary and Janelle1 Also, antibiotic resistance is a growing concern throughout healthcare. 2,Reference Marston, Dixon, Knisely, Palmore and Fauci3 Studies on de-escalation of antibiotic therapy in HAP have shown outcomes to be no worse for patients who had antibiotics de-escalated compared with those who did not. Reference Ilges, Ritchie and Krekel4–Reference Masterton6 Therefore, it is prudent to further characterize existing rates of antibiotic de-escalation among HAP patients. In this study, we sought to determine whether any systematic differences in de-escalation patterns exist between 4 pathogen groups (ie, Enterobacterales, Staphylococcus aureus, Pseudomonas aeruginosa, and bacteria-culture–negative status) in patients with HAP and ventilator-acquired pneumonia (VAP) pneumonia.
Methods
The methods used to create this database have been described previously. Reference Ilges, Ritchie and Krekel4 This retrospective cohort comprised patients was admitted to Barnes Jewish Hospital, a large academic center in St. Louis, Missouri, between January 16, 2016, and December 31, 2019. The study was approved by the Washington University institutional review board. Adult patients with ICD-9 or ICD-10 codes and who met the following criteria for HAP or VAP were included: (1) index date of infection onset ≥48 hours after hospital admission for HAP or ≥48 hours after initiation of mechanical ventilation for VAP, (2) chest radiograph completed within ±24 hours of index pneumonia date, (3) active orders for either anti–methicillin-resistant Staphylococcus aureus (MRSA) or anti–Pseudomonas aeruginosa antibiotics, and (4) at least 1 sign of infection, including a white blood cell count ≥11 or ≤4×109 cells/L or temperature ≥38 or ≤36°C. The index date of pneumonia diagnosis was defined as day 0. Pharmacy-verified intravenous and oral antibiotic orders of interest were collected for each day of admission.
Antibiotic spectrum scores modified based on the Barnes-Jewish Hospital antibiogram and the addition of antibiotics not listed in the original description reported by Gerber et al Reference Gerber, Hersh, Kronman, Newland, Ross and Metjian7 were calculated on day 0 and for each day thereafter. Reference Ilges, Ritchie and Krekel4,Reference Gerber, Hersh, Kronman, Newland, Ross and Metjian7 Microbiologic data, including respiratory cultures, respiratory viral polymerase chain reaction tests, MRSA nasal culture swabs, and blood cultures, were collected from days –1 to 3 of index pneumonia diagnosis. Respiratory cultures were classified as sputum-like specimens, which included sputum and tracheal aspirates, and bronchoscopy specimens, which included bronchoalveolar lavage, washings, and brushings. Respiratory samples identified as positive for a likely pathogen excluded those with only yeast, fungal structural elements, and/or clinically insignificant flora. Among the blood cultures identified as positive for a likely pathogen, we excluded those with only coagulase-negative staphylococci. Culture negative was defined as no growth from any of the cultures described above.
Study participants were categorized in 4 groups based on the pathogen associated with their pneumonia: Staphylococcus aureus (n = 134), Pseudomonas aeruginosa (n = 59), Enterobacterales spp (n = 87), and bacteria-culture negative (n = 1,404). Due to small sample sizes, study participants with pathogens not of interest were excluded from analysis. These pathogens included Stenotrophomonas, Burkholderia, and Acinetobacter as well as S. pneumoniae, H. influenzae, and S. agalactiae.
The spectrum scores from each pathogen group were compared separately on days 0, 3, 7, 10, and 14 using Kruskal-Wallace tests and subsequent Dunn post-hoc tests if warranted. The initial spectrum scores were calculated on the day study participants had a new active order for either anti-MRSA or anti–Pseudomonas aeruginosa antibiotics and at meeting the study criteria for HAP and VAP described above. The composite outcome of all-cause mortality and hospital readmission within 30 days of pneumonia diagnosis was also calculated similar to our prior study. Reference Ilges, Ritchie and Krekel4 All analyses were performed using R version 4.1.3 and R Studio version 2022.02.3 (R Foundation for Statistical Computing, Vienna, Austria).
Results
The demographic characteristics of this cohort have been described previously. Reference Ilges, Ritchie and Krekel4 In total, 11,860 admissions were screened for inclusion, and 1,684 of these met inclusion criteria for this study. The final cohort comprised 87 patients (5.2%) with Enterobacterales infections, 134 (7.9%) with Staphylococcus aureus infections, 59 (3.5%) with Pseudomonas aeruginosa infections. In addition, 1,404 (83.4%) had bacteria-culture–negative pneumonia. The median age was 62 years (interquartile range [IQR], 51–70.8). In total, 975 study participants (57.9%) were male, and 1,231 (73.1%) were white. The median Charlson comorbidity index score was 6 (IQR, 3.8–8). Overall, 1,057 patients (62.8%) were diagnosed with HAP and 627 patients (37.2%) were diagnosed with VAP. 1483 (88.1%) patients were in the ICU on the day of diagnosis.
In comparing spectrum scores between pathogen groups on days 0, 3, 7, 10, and 14, only day 0 showed a statistical difference in spectrum scores between groups (Kruskal-Wallace test P < .0001) (Fig. 1). On this day, the bacteria culture-negative group had a higher spectrum score than the Enterobacterales and Stapylococcus aureus groups (Dunn post-hoc test P < .001). None of the other analyzed days showed statistically significant differences in spectrum scores between groups (Kruskall-Wallace test P > .10) (Fig. 1). A significantly higher percentage of patients with positive bacterial cultures (70.3%) than bacteria-culture–negative patients (56.1%) remained on antibiotics at day 7 (χ2 test P < .001).
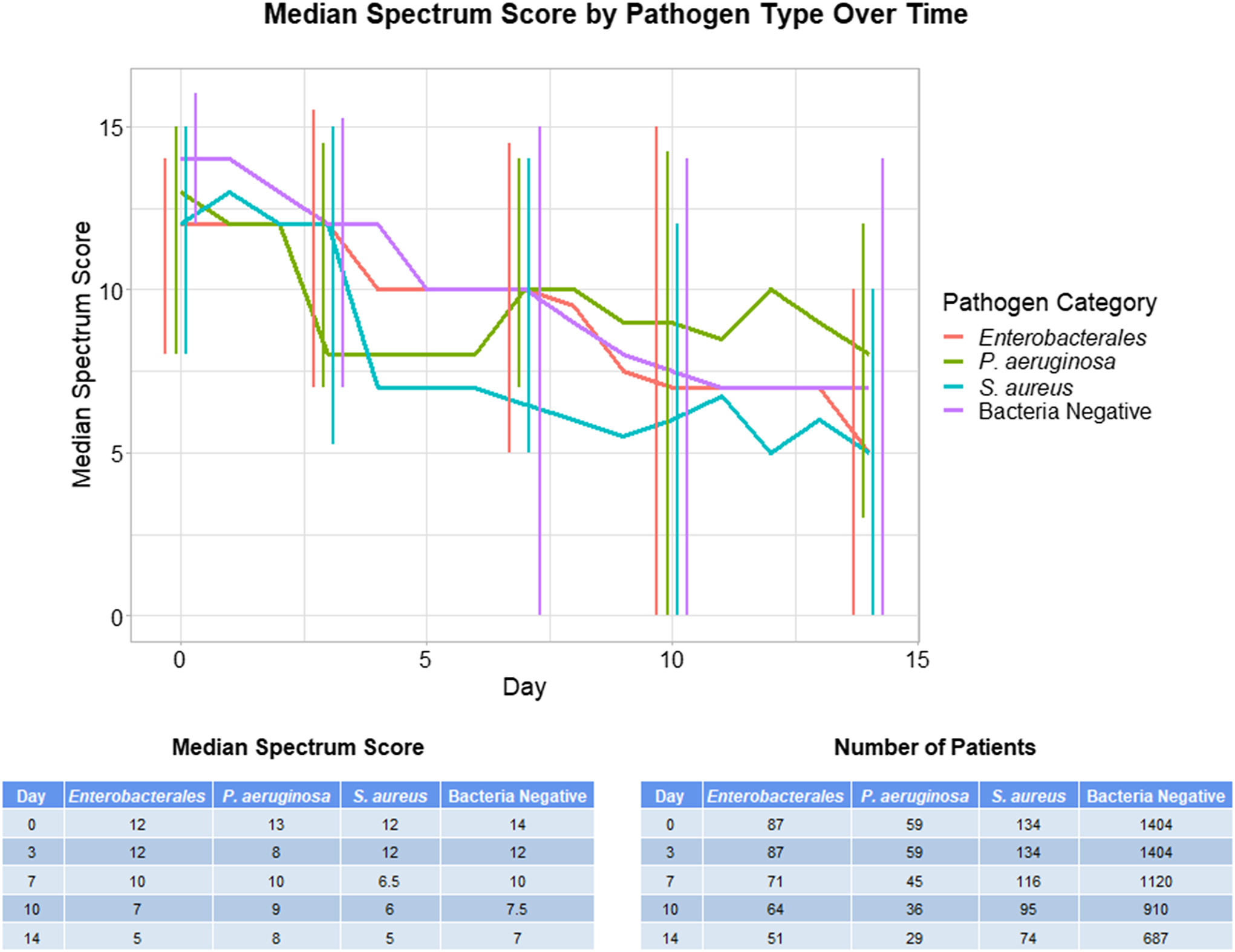
Fig. 1. Median antibiotic spectrum scores over time on days 0, 3, 7, 10, and 14. Error bars indicate interquartile range.
The pathogen groups exhibited outcomes that were not significantly different based on a composite of all-cause mortality and hospital readmission within 30 days of pneumonia diagnosis (χ2 test P = .049; χ 2 post-hoc test P > .10). Moreover, 26.1% of patients in the Staphylococcus aureus group, 34.8% in the bacteria-culture–negative group, 40.2% in the Enterobacterales group, and 44.1% in the Pseudomonas aeruginosa group reached the composite end point.
Discussion
The result of this retrospective cohort study provide no evidence that antibiotic de-escalation occurs at different rates based on the pathogen identified to be causative of HAP or VAP. This information is important because differences in treatment patterns between pathogen groups would have merited further study. Along with previous studies indicating that patient outcomes are no worse when antibiotic therapy is de-escalated, our data indicate that opportunities for further de-escalation can be pursued based on microbiologic pathogen type. Reference Ilges, Ritchie and Krekel4–Reference Masterton6 Most interestingly, bacteria-culture–negative patients did not have lower rates of de-escalation compared to patients with an identified bacterial pathogen.
Our study had several limitations. The study included the small number of organisms recovered, predisposing to a type 2 error in de-escalation rates between groups and the inherent difference in antibiotic spectrum based on positive cultures driving de-escalation. That is, stopping linezolid versus meropenem when MRSA or ESBL gram-negative organisms, respectively, were not identified. Additionally, it is possible more patients in the culture negative group did not have pneumonia. This diagnostic uncertainty may have contributed to continued empiric antibiotic therapy, impeding any attempt at de-escalation. Finally, there are potential limitations of evaluating antibiotic spectrum rather than a broader off-target impact of antibiotics on microbiota which may more directly influence antibiotic resistance emergence and outcome. Reference Ruppé, Burdet and Grall8
Antibiotic stewardship remains a challenging issue in the present time, especially among patients facing life-threatening conditions or those in ICUs. A recent cluster-randomized trial from a large US academic center found that weekly multidisciplinary antimicrobial stewardship round making was a high-resource intervention associated with a small impact on antibiotic utilization. Reference Seidelman and Turner Na9 Another recent study from Singapore found that an intensive antimicrobial stewardship program employing prospective review and computerized decision support systems was associated with limiting the use of pipercillin-tazobactam and carbapenems while reducing use of other antibiotics as well. Reference Ng, Heng and Chua10 Our findings suggest opportunities for further de-escalation that can occur based on quantitative assessments of antibiotic spectrum, especially for patients with bacteria-culture–negative HAP and VAP.
Acknowledgments
Financial support
Dr Kollef’s efforts are supported by the Barnes-Jewish Hospital Foundation.
Conflict of interest
All authors have no conflicts of interest to report.



