Introduction
Human papillomaviruses (HPVs) are agents of the most common sexually transmitted diseases. The approximately 45 HPV genotypes that infect the genital mucosa are classified as low-risk HPV (LR-HPV) or high-risk HPV (HR-HPV) on the basis of their association with premalignant and malignant lesions [Reference Egawa1]. HR-HPV plays a key role in the development of cervical cancer, which is the fourth most common cancer in women worldwide (the second in low socio-demographic index countries) and in a variable proportion of non-cervical malignancies, including cervical squamous cell carcinoma [Reference Syrjanen2] and cancer of the penis [3].
HPV infection in men shows a prevalence varying from 1.3% to 72.9% with a peak of 65.4% between 18 and 40 years [Reference Capra4]. Since the presence of the virus has been widely reported in semen, there is a growing interest in the impact of HPV infection on male fertility and reproductive health [Reference Foresta5]. A recent work by Lyu et al. [Reference Lyu6] estimated the prevalence of HPV in semen to about 11% in men from general population and even 20% in fertility clinic attendees. Recent results demonstrated a possible correlation between HPV sperm infection and cases of idiopathic asthenozoospermia and unexplained male infertility but literature data are still controversial [Reference Foresta7–Reference Foresta9], and the mechanism by which HPV could affect sperm motility is still unknown.
Likewise, the exact HPV localisation in the different components of semen as well as the role of infected sperm cells as transmission vectors for the virus are still unclear. Literature analysis revealed that HPV DNA is generally located on sperm surface by binding efficiently to two sites along the equatorial region of the sperm head [Reference Schillaci8]. This binding would involve the capsid protein L1 and the syndecan-1 receptor [Reference Foresta9].
However, the presence of HPV DNA inside the sperm cells and its physical state is still investigated. Two important questions must be answered. Can HPV DNA enter into the sperm cell? It is known that in animal models spermatozoa have the intrinsic ability to bind and internalise exogenous DNA during their transfer from testis to penis (through the vas deferens) and to transfer it into the egg at fertilisation [Reference Kojima10]. What is the HPV DNA physical state in sperms? It is well known that, in the late stages of most cervical cancers in women, HPV genome can recombine with chromosomal DNA and this event can yield integration in the cellular genome [Reference Wentzensen, Vinokurova and von Knebel Doeberitz11, Reference Bosco12].
In order to shed some light on HPV infection in semen, we examined samples from partners of women with symptomatic HPV infection (high-grade squamous intraepithelial lesion (HSIL)). From each subject, seminal parameters were evaluated. HPV was detected and genotyped by conventional protocols. Moreover, HPV DNA localisation in the different semen components was detected by means of a new protocol that we developed. By genotype-specific qPCR, we also determined the viral DNA quantity of different HPV types in spermatozoa and virus effect was evaluated in the isolated semen fractions. HR-HPV DNA integrity was also studied.
Materials and methods
Patient enrolment and sample collection
Male partners of women with HSIL-positive for HR-HPV, who were treated at the Clinic of Obstetrics and Gynecology of the University of Palermo, consented to their inclusion in a prospective observational study from January 2016 to December 2016. All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the local Ethics Committee (Project identification code: B71J11000160007). All patients who met the following inclusion criteria were enrolled: age between 18 and 50 years; partner positive for HR-HPV types; sperm concentration after swim up procedure that exceeded 1 million/ml.
Exclusion criteria were: varicocele; cryptorchidism; other genital infections; patients treated with different medical therapies (chemo/radio therapy).
Semen processing
Semen samples were obtained by masturbation after 3–5 days of sexual abstinence. After liquefaction at room temperature, semen volume, sperm concentration, motility and normal morphology were evaluated according to World Health Organization guidelines [Reference Björndahl13] for semen analyses. Sperm cells were then separated by swim-up technique. This technique allows to select the spermatozoa for their ability to migrate from seminal plasma to the culture medium. After centrifugation at 300 g for 7 min, supernatants were discarded and 1 ml of Sperm Preparation Medium (Origio-Medicult, Sweden) was gently stratified on the pellet. Samples were then incubated at 37 °C in 5% CO2 for 45 min, then a sterile Pasteur pipette was used to collect the supernatant containing actively motile sperms.
An aliquot of the total semen sample and an aliquot of semen obtained by the swim-up procedure (SU fraction) of each patient were sent to the Virology Laboratory for HPV detection and genotyping.
Other two aliquots of total semen and SU fraction, respectively, were sent to the Molecular Biology Laboratory for virus research in the various components of the seminal sample and to evaluate the physical state of viral DNA.
Detection and genotyping of HPV DNA
For detection and genotyping of HPV, DNA was extracted from total semen and SU fraction by the QIAamp Mini Kit (Qiagen).
Ten microliters of DNA were used for HPV genotyping by INNO-LiPA HPV Genotyping Extra (Fujirebio Europe), which is based on the combined use of a PCR and a reverse hybridisation assay. The amplification was performed according to the manufacturer's instructions. After PCR, denatured amplicons were hybridised with specific probes for 28 HPV genotypes. The assay covers all currently known HRHPV genotypes and probable HR HPV genotypes (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70,73, 82) as well as a number of LR-HPV genotypes (6, 11, 40, 43, 44, 54, 70) and some additional types (69/71, 74).
Differential DNA extraction
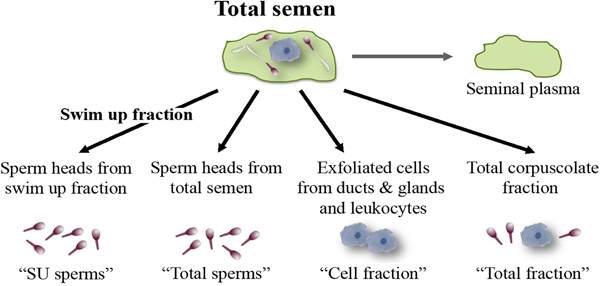
Total semen and SU fraction from each patient were processed in order to carry out a differential lysis with consequent DNA extraction from the semen components separately (Fig. 1). To this purpose, we used the Blood and Cell Culture DNA kit (QIAGEN) according to manufacturer's instructions with some modifications. During the tuning phase of the protocol, at every step, samples were observed under microscope to verify cell and sperm lysis (Fig. 2). Samples were washed twice with PBS obtaining a total corpuscolate fraction (Total fraction) from total semen. From Total fraction, DNA was extracted as control. Cell Lysis buffer (1.28 M Sucrose, 40 mM Tris-HCl pH 7.5, 20 mM MgCl2, 4% Triton X-100) was used to break the membranes of all cellular components except for the sperm membranes. Instead, a Digestion buffer (800 mM Guanidine-HCl, 30 mM Tris-HCl pH 8.0, 30 mM EDTA pH 8.0, 5% Tween-20, 0.5% Triton X-100) was used to break the nuclear membranes of previously lysed cells and to detach sperm tails. This buffer does not break sperm heads (Fig. 2a).
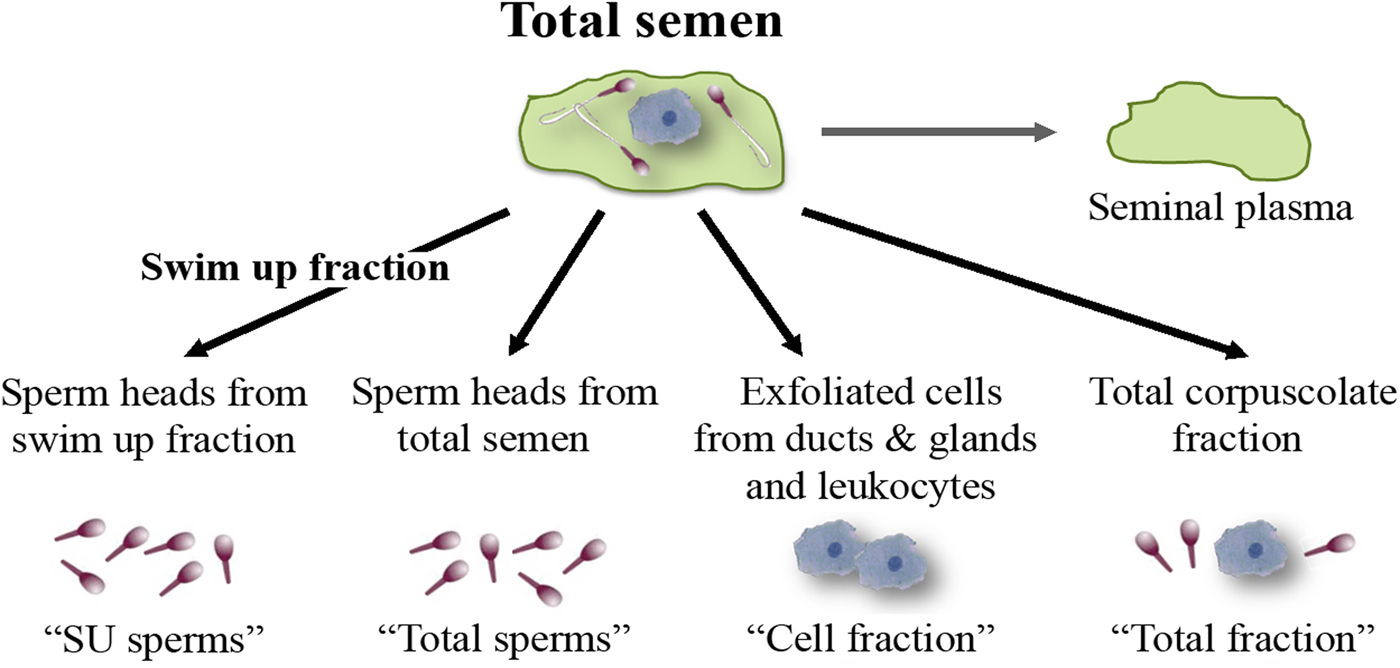
Fig. 1. Semen fractions for differential DNA extraction.
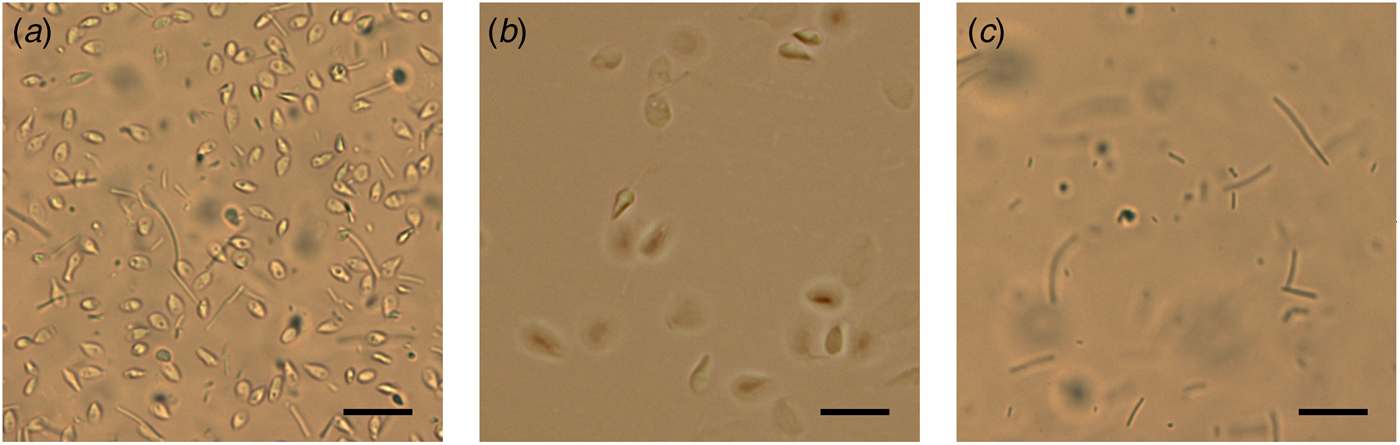
Fig. 2. Microscope observations during differential lysis procedure. (a) Spermatozoa heads and detached tails after incubation in Digestion buffer. (b) Washed sperm heads in Digestion buffer without Dithiothreitol. (c) Unwashed control: sperm tails after incubation with Digestion buffer containing Dithiothreitol and Proteinase K. Scale bar: 25 µm.
After centrifugation at 1500 g for 5 min, the supernatants containing the lysate cells (Cell fraction) were unified. The two pellets containing only sperm heads from total fraction (Total sperms) and from Swim Up fraction (SU sperms) were washed twice with the Digestion buffer (Fig. 2b). Pellets were resuspended in a Digestion buffer containing Dithiothreitol (DTT 0.5 mM), to lyse sperm heads. All the fractions were incubated at 50 °C for 60 min with Proteinase K (>15 mAU/ml). Subsequently, DNA purification was performed by anion exchange chromatography following the instructions provided by the kit. DNA purity and concentration were assessed via spectrophotometry; DNA integrity was assessed on agarose gel.
Viral DNA detection in the different semen fractions
The search for HPV DNA in the various semen fractions was performed by nested PCR with the use of universal primers (MY09/11, GP5+/6+). The initial outer PCR was carried out using the following conditions: 100 ng DNA 94 °C for 5 min followed by four cycles at 94 °C for 60 s, 45 °C for 60 s and 72 °C for 60 s, followed by 30 cycles at 94 °C for 30 s, 45 °C for 30 s and 72 °C for 30 s and a final cycle at 72 °C for 5 min. Nested PCR: one microliter of the first round product was used for the second amplification using the following parameters: 94 °C for 2 min followed by 30 cycles at 94 °C for 30 s, 45 °C for 30 s, 72 °C for 20 s and finally, one cycle at 72 °C for 5 min. A HeLa cell line DNA was used as the positive control and no template DNA as the negative control. Moreover, as internal control, β-globin locus (HBB) primers (GH20/PCO4) were also used.
Real-time PCR for viral DNA quantification and genome integrity evaluation
Real-time PCRs were performed in 96-well plates in a 20 µl mixture containing 200 ng of the sperm DNA (Total sperm fraction), 10 µl QuantiFast SYBR Green PCR mix (Qiagen) and 1 µM of each primer, in the BIO-RAD CFX96 system using the following PCR parameters: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s. Following amplifications, melt curves were generated by heating from 65 to 95 °C with 0.5 °C increments and 5 s dwell time [Reference Ragusa14].
In order to test the HR-HPV genome wholeness, six different regions were tested: E1n (the region coding for the N-terminus of E1 protein), E1c (the region coding for the C-terminus of E1 protein), E2, L2, L1 and LCR (Long Control Region). The sequences of the specific primer pairs used for qPCR are shown in Table 1.
Table 1. List of primers

Target genes, primer names (numbers designate the position of the 5′ end with respect to the GenBank reference sequences (AY262282 for HPV18 sequence)) and primer sequences used in qPCR experiments. Sizes of the amplicons are also indicated.
Real-time PCR experiments were repeated once, each time every target was run in triplicate, with negative controls (without DNA) and positive controls (25 ng HeLa cervical carcinoma cell DNA). The absence of non-specific products was confirmed by both the analysis of the melt curves and electrophoresis in 2% agarose gels. Amplification efficiency of each primer pair was previously evaluated by serial dilutions of DNA.
HBB (β-globin) gene was used as internal reference (GH20/PCO4 primers) and the gene copy numbers were analysed by using the CFX Manager software. Viral DNA quantity was presented as HPV copies/sperm as determined by the viral LCR copies per reaction/HBB copies per reaction. The integrity status of HPV was assessed using a ratio of the tested gene copies (E1n, E1c, E2, L2, L1) over the LCR copies per reaction.
Statistical analysis
Results were expressed as a mean value ± s.d. Real-time experiments were repeated once, each time every target was run in triplicate. All data were statistically analysed using SPSS software for Windows. P values <0.05 were considered statistically significant and were indicated by ∗. P values lower than 0.005 were indicates by **, P values lower than 0.0005 were indicated by ***.
Results
Seminal parameters, HPV detection and genotyping
Semen samples of 22 patients aged between 22 and 44 years (mean age 32 years), partners of HPV-positive women with HSIL were analysed for semen volume, sperm concentration, motility and morphology. Sperm concentration was determined also after swim up procedure (Table 2).
Table 2. Semen parameter evaluation, HPV detection and genotyping results

PMS, progressively motile sperm; NPMS, non progressively motile sperm; NMS, non motile sperm.
HPV detection was performed by the conventional protocol (left) and by the new method: after differential lysis, DNA was extracted from separate fractions and HPV detection was performed by nested PCR (right columns). Samples were ordered based on HPV positivity. Samples resulted positive by HPV conventional detection are in bold.
The HPV infection was evaluated both in total semen and in SU fraction. Forty-five per cent (10/22) of patients had the infection in the semen sample (Total semen). HPV test was positive in three samples also after swim up technique (SU fraction).
HPV genotyping showed that all positive samples contained at least one high-risk genotype and there was a prevalence of HPV16 and HPV18 (6/10 = 60%). Among the 10 infected couples with an HPV test positive in total semen, 60% (6/10) showed the concordance of at least one genotype and in each case it was HR-HPV.
Presence of HPV DNA in different semen components
DNA was extracted after differential lysis from different semen fractions: washed spermatozoa (Total sperms), spermatozoa from swim up procedure (SU sperm), somatic cells (Cell fraction) and corpuscolate fraction (Total fraction) as control. Nested PCRs, performed on the different semen fractions using universal HPV primers, revealed differences between samples. Results reported in Table 2 show that the viral DNA can be detected in every fraction of semen: Total sperms, Cell fraction and seminal plasma. Differently with respect to genotyping results, by this method we never found HPV DNA in the sperm heads recovered after the swim up and the differential lysis procedures (SU sperms). Different samples can contain HPV DNA in both Total sperm and Cell fractions or just in one of them. Moreover, three samples contained viral DNA only in the seminal plasma. A sample resulted positive in the ‘total fraction’, but negative in the separated fractions. That can seem an inconsistency, however, when the virus is in the liquid fraction of the semen, it could adhere to cells/sperms, but it would be lost after the hard washes (Cell Lysis buffer and Digestion buffer) required for the differential extraction.
Four samples resulted containing viral DNA in the sperms. One of them (sample 1) was infected by three different HPV genotypes: HPV31, classified as HR carcinogen, HPV73, classified as HR non carcinogen and HPV44, classified as LR mucosal. Sample 3 contained HPV73 (HR non-carcinogen) and two samples (2 and 4) presented the genotype HPV18 wich is involved in the majority of HPV-related cancers toghether with HPV16 [Reference Serrano15].
Type specific viral DNA quantification
In order to study the infection in samples resulted positive for HPV in spermatozoa, we assessed the viral DNA quantity with respect to human genome copies. Viral DNA ratio evaluation was carried out by real-time PCR using primers specific for each HPV genotype and corresponding to LCR, using β-globin locus (HBB) as reference.
Quantification of the LCR-HPV/HBB ratio allowed the estimation of viral copy number ratio. As control, the same reactions were performed on DNA extracted from HeLa cervical carcinoma cell line, which contains several copies of HPV18 DNA. Results obtained for HeLa cells showed that the LCR/HBB ratio was 14 (±3). Since HeLa cells are hyper-triploid and have three HBB locus copies, results indicated approximately 40 copies of HPV18 per cell, according to literature data [Reference Adey16].
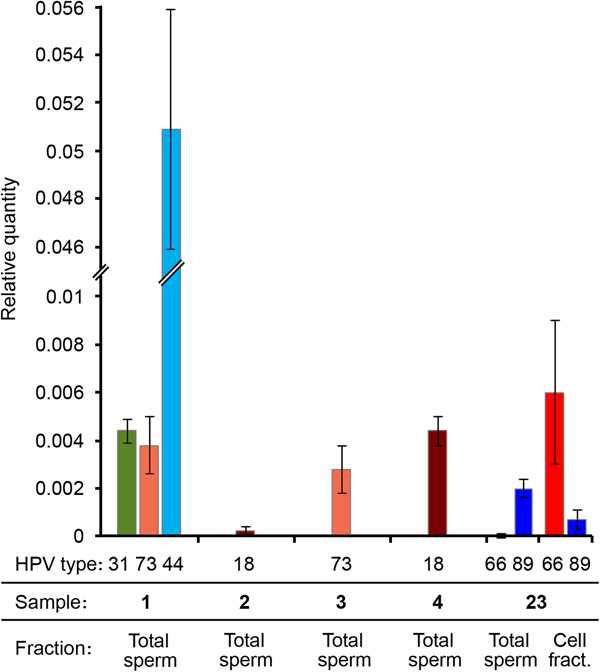
RT-PCR results showed that in all the tested samples the LCR/HBB ratio was very low for the HR-HPV genotypes (approximately 0.4%). The LCR Ct value for the sample 2 was too high (>35), therefore the viral quantity was even lower and unassessable. Therefore, these data indicated a low viral DNA ratio for all samples infected by a HR-HPV (31, 73, 18). Interestingly, the viral DNA ratio of the LR mucosal HPV (HPV44) was 10 times higher (5%) (Fig. 3). It is worth noting that samples showing higher viral DNA ratio had lower percentage of progressively motile sperms (Table 2).
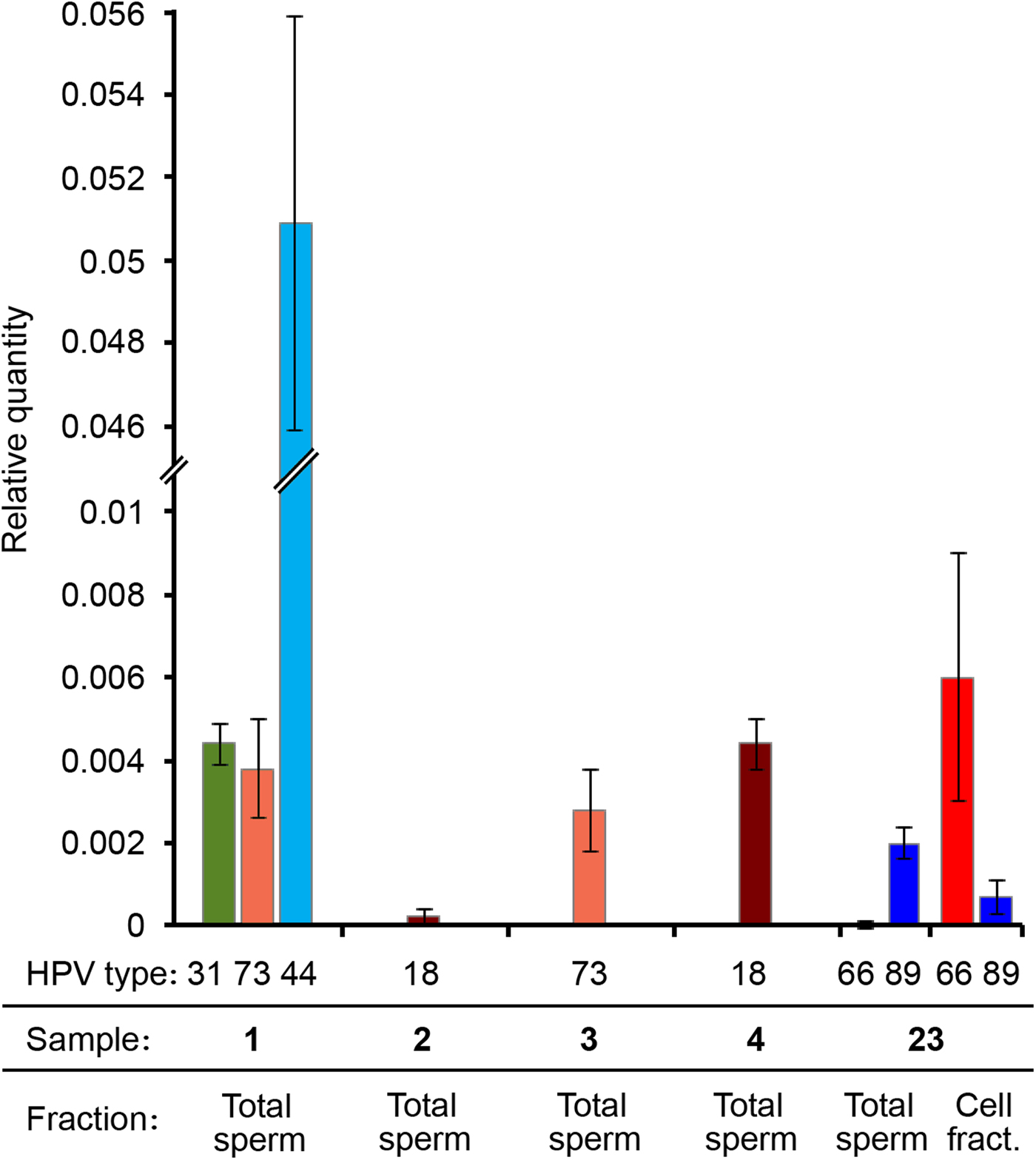
Fig. 3. HPV viral DNA ratio determined by real-time PCR. Different HPV genotypes, sample numbers and fractions are indicated. Viral DNA quantity is expressed as relative quantity with respect to β-globin locus. Total sperm: DNA extracted from the washed sperm heads derived from the total semen. Cell fract.: DNA extracted from the somatic cell fraction.
We performed qPCRs also on another sample (number 23) genotyped as HPV66 and HPV89, resulted positive both in spermatozoa and in somatic cells by nested PCR and that was not included in the study because seminal parameters were not determined. Surprisingly, in this sample HPV89 DNA was present only in the Total sperm fraction, whereas HPV66 was principally detected in the cell fraction (Fig. 3).
HR carcinogen HPV genome integrity evaluation
Since HR carcinogen HPV, in particular HPV18 and 16, can recombine in cervical cells and those events can elicit viral genome integration [Reference zur Hausen17], we decided to analyse the physical state of HR carcinogen HPV found in spermatozoa. An analysis of the wholeness of the viral genome was performed by real-time PCR, using primers corresponding to different regions of the viral genome. Indeed, the ratio between the copy number of different HPV regions allows estimation of viral integrity [Reference Cañadas18].
Six different regions were tested (Table 1 and Fig. 4b). Since LCR is essential for HPV replication and transcription, this region was selected as HPV reference.

Fig. 4. Inspection of viral genome integrity. (a) Results of real-time PCRs performed on HPV18 DNA extracted from a sample resulted positive in total spermatozoa (sample 4) to determine the ratio between selected HPV regions and LCR. Reference gene was HBB locus. Data points represent means (three replicates for each target and experiments were carried out two times) with standard deviations. **P values <0.005. (b) From inside to the outside of the circle: (1) The circular dsDNA genome of HPV18 is shown. Viral open reading frames are depicted as orange arrows. Transcription and replication control region (LCR) is depicted as a blue rectangle. (2) RT-PCR amplicon locations are indicated. (3) Putative part of the viral genome deleted in sperms is also indicated in red.
RT-PCR results performed on sample 1 (HPV31) showed that the copy number of all tested regions were approximately the same.
Results obtained on sample 4 (HPV18) showed that the copy number of L1 and L2 was approximately equal to LCR copy number. Differently, E1n, E1c and E2 copy number ratio was lower than 0.5 with respect to LCR (Fig. 4a). These data indicate that, in this sample, less than half viral genomes are complete and more than half are defective. These results suggest a possible recombination event during which a disruption of the HPV genome occurred, causing the deletion of E1 and E2 genes (Fig. 4a). The copy number ratios suggest that not every sperm in this sample contains viral DNA and that both complete and defective forms are present, resulting in a mixed state.
Discussion
In concordance with literature data, results from this study showed a high prevalence of HPV infection in male partners of women with HSIL [Reference Partridge and Koutsky19]. In most cases, infection was caused by high-risk genotypes, with a prevalence of HPV16 and 18. This confirmed once again the close association between persistent infection in women and viral genotypes with high oncogenic potential, resulting in an increased risk of neoplastic evolution of the injury [Reference Capra20]. These results also confirmed that the sexual contact is the main way of transmission of HPV infection and the concordance of at least one viral genotype, in each case at high risk, in the majority of couples would confirm the hypothesis, already advanced in the recent literature [Reference López Diez21, Reference Giovannelli22], of an important role of infected male in the transmission and/or maintenance of infection in women with an increased risk of cervical cancer. In this study, for the first time, we applied a protocol of differential extraction from semen achieved by differential lysis of epithelial cells and sperm in the absence and presence of DTT that is used also in forensic biology protocols for forensic STR analysis when DNA from epithelial cells (of victim) and sperm (from perpetrator) in samples from sexual assault have to be isolated separately for obtaining independent genotypes [Reference Lee and Shewale23]. The differential lysis method applied for the first time in this study allowed us to inspect thoroughly semen infection, revealing that all the semen components (spermatozoa, somatic cells and/or plasma) can contain viral DNA. This finding is in concordance with other literature data [Reference Foresta7] and with the recent detection of HPV16 infection markers in CD20+ and CD56+ leukocytes of patients with HPV16 infection in semen reflecting the presence of virus particles within the endosomal compartment [Reference Foresta24] and a mechanism for the viral binding to any cell has been proposed [Reference Surviladze, Dziduszko and Ozbun25]. Semen infections are heterogeneous: different samples can contain the virus in different fractions and more than one HPV genotype can be found in the same fraction. Moreover, in the event that multiple HPV genotype infection occurs in the same sample, co-infecting genotypes can be detected in different semen fractions. Unfortunately, no correlation between HPV genotypes and different virus localisation was found in these samples. Indeed, for example, HPV18/16-positive semen samples had the virus in every kind of fraction combination (only somatic cell, only sperm, both cell and sperm fractions, etc.). Interestingly, the sole LR mucosal HPV found in spermatozoa fraction (HPV44) showed a DNA copy number 10 times higher than the overall burdens observed in HR-HPV infected samples. HPV44 belongs to the α10 species and is associated with condylomata and low-grade cervical changes [Reference Capra26], and causes anogenital warts [Reference Aubin27]. The occasional role of this LRHPV in anogenital carcinoma has been demonstrated [Reference Guimerà28] and HPV44 has been detected also in stool samples [Reference Di Bonito29].
The heterogeneous virus distribution supports the assumption that a variety of factors and mechanisms could be involved in transporting HPV DNA. Consequently, many possible modes of sexual transmission of HPV may exist, explaining also the conflicting results existing in the literature about HPV infection in men. The hypothesis of a possible correlation between HPV type and mechanism(s) involved in transmission of infection could be the subject of a further work.
Besides, the effect of HPV presence in semen is extensively debated and the presence of the virus has been associated with the impairment of sperm motility [Reference Lai30], suggesting that HPV infection could be an emerging risk factor for male infertility. It is noteworthy to consider that the effect of HPV DNA on semen could be genotype-dependent [Reference Hiller31]. We have previously reported [Reference Schillaci8] no effect of HPV infection on sperm parameters when we compared HPV-positive total semen samples with respect to HPV-negative counterparts. In this study, the new method of inspection opens the possibility to easily distinguish different semen infections and to observe possible effects on semen.
It is worth noting that the results obtained by the two virus detection methods were different for SU fraction (total DNA extraction and reverse hybridisation) and SU sperms (DNA extraction from washed sperm heads and nested PCR), indicating a different sensitivity of the used techniques or the inability of the swim up procedure to eliminate virion contamination and as a consequence actually no infected sperms are present in the SU fraction, indicating also that infected sperms cannot swim normally.
Our study confirms the possibility of the existence of a strong binding between virus and spermatozoa already demonstrated by Foresta et al. [Reference Foresta9] as a matter of fact we found the presence of HPV DNA in the sperm fraction previously washed with a solution containing two strong chaotropic agents, a surfactant and a detergent. All that would indicate the presence of HPV DNA into the sperm (at least in some sperms, as the low viral quantity observed) and not only a simple external interaction between virus and sperm. Even though viral multiplication in spermatozoa seems to be unlikely because the life cycle of HPVs is dependent on the cell cycle progression into mitosis and on the differentiation of basal cells [Reference Graham32], sperm cell infection may be a mechanism for a direct delivery of HPV DNA in the oocyte [Reference Foresta9]. Inside the zygote, viral genes, previously silenced by the chromatin compaction in sperm, could be activated and transcribed so influencing cell cycle and development regulation. In fact, it has been recently shown that HPV detection on sperms was predictive of negative pregnancy outcome [Reference Garolla33].
Our results also suggest a possible recombination event in half of viral genomes in a HPV18 sample infected in spermatozoa, with a deletion of E1 and E2 genes. Since recombination events in cervical cancer are related to HPV integration in the host genome [Reference Wentzensen, Vinokurova and von Knebel Doeberitz11], it will be interesting to examine in depth this possibility. In any case, integrated or not, it will be also interesting to know when, where and how viral DNA can enter the sperm: during spermiogenesis in the seminiferous tubules or after spermiation, during epididymal maturation or when the sperm travels through the vas deferens. It is in fact known that in men infections can spread via urethra, prostate gland, seminal vesicles, deferent duct, epididymis until testis like in male accessory gland infection [Reference Krause34].
It was important that none of the infected males showed visible lesions, in contrast to several studies where it was reported that the penile lesions were much more frequent in partners with high-risk genotypes reported by Patridge and Koutsky [Reference Partridge and Koutsky19]. On the other hand, Luttmer et al. [Reference Luttmer35] recently showed an association between HPV infection of the penile epithelium and viral DNA presence in semen of healthy men, suggesting that virus can spread in semen by the desquamation of the penile cells.
The presence of HPV DNA even in the semen of normospermic patients suggests the need of screening for HPV infection in all patients who undergo assisted reproduction techniques [Reference Ruvolo36].
Also, since it seems certain that the sexual behaviour of males may contribute to an increase in the risk of cervical cancer in their partners [Reference Castellsagué37] and more than a quarter of the HPV-associated cancers in American males affects the oropharyngeal tract and anorectal tracts [Reference Muñoz38], the ‘Center for Control and Prevention of Disease’ (CDC) recommends vaccination of men at the same level of vaccination for women [Reference Markowitz39]. Also healthy men can carry a range of high-risk and/or low-risk types of HPV genotypes in the genital area [Reference Yin40]; therefore, the extension of vaccination to sexually active males would lead to a significant reduction of the transmission of the infection and to a reduction of the costs of therapeutic treatment of the associated lesions [Reference Sheikh41].
This work demonstrates that the new protocol is useful to study HPV infection thoroughly and can open the way to new epidemiological studies. We will continue to collect and study other samples, in order to increase the sample size and the significance of our results.
Author ORCIDs
G. Capra, 0000-0002-1407-7282; M. A. Ragusa, 0000-0002-8760-1022
Acknowledgements
We would like to thank Professor Fabrizio Gianguzza for his valuable suggestions and critical reading of the manuscript. We thank also Professor Aldo Di Leonardo for providing HeLa cell line.
Author contributions
GC, RS and MAR performed the experiments and collected the data; LB, MCR and AP conceived the study and revised the manuscript; RS drafted the manuscript; GC and RM took part in critical revision and manuscript editing; GC, RS and AP provided the patients; AP provided the financial support for the project; MAR analysed the data, made figures and tables and drafted, reviewed and revised the manuscript. All authors have read and approved the final version of the manuscript, and agree with the order of presentation of the authors.
Financial support
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest
None.