Acute respiratory infections (ARI) are prevalent worldwide and directly responsible for morbidity and a significant proportion of mortality, mainly in children. The most frequent agents of ARI are respiratory viruses, including human respiratory syncytial virus (HRSV), human metapneumovirus (HMPV), human rhinovirus (HRV), human influenza viruses A and B (Flu A, Flu B), human parainfluenza viruses (HPIV) 1, 2 and 3, human coronaviruses (HCoV) and human adenoviruses (HAdV) [Reference Arruda, Cintra, Hayden, Guerrant, Walker and Weller1]. Recently, a parvovirus named human bocavirus (HBoV) was discovered in respiratory specimens [Reference Allander2]. Since then, HBoV has been detected in different geographic regions, suggesting a worldwide distribution. In order to assess the frequency of HBoV in two hospitals in Ribeirão Preto, SP, Brazil, respiratory samples from children admitted with ARI in 2005 were tested by polymerase chain reaction (PCR).
Nasopharyngeal aspirates (NPA) were collected in 2005 (January–December) from children aged <5 years of age, who were admitted with ARI to either the University of São Paulo Hospital or Santa Lydia Hospital, both located in the city of Ribeirão Preto, state of São Paulo, southeast Brazil. The samples were obtained as previously described [Reference Rakes3] with minor modifications. Briefly, 2 ml physiological saline was instilled into each child's nostril, followed by aspiration of the wash fluid and secretion from both nostrils into a sterile collection trap by gentle suction. The NPA was transferred to a 15 ml conic tube and the final volume was completed to 3 ml with phosphate-buffered saline (PBS, pH 7·2; Gibco/Invitrogen, Carlsbad, CA, USA). Each sample was treated for 1 h at 4°C with 10% (w/v) antibiotic-antimycotic solution (30 U penicillin G, 30 μg streptomycin and 0·075 μg amphotericin-B in 0·85% PBS; Gibco) plus 30 μg ciprofloxaxin (Bayer, Leverkusen, Germany). NPA were then were aliquoted, including one sample with 250 μl NPA plus 750 μl TRIzol® (Invitrogen, Carlsbad, CA, USA), which was kept at −70°C for nucleic acid extraction. From that aliquot, total RNA was extracted using the manufacturer's protocol and a DNA Purification kit (Promega, Madison, WI, USA) was used for DNA extraction from the DNA-enriched Trizol fraction.
PCR for HBoV was performed using Taq DNA polymerase (Biotools, São Paulo, Brazil), primers 188F (5′-GAGCTCTGTAAGTACTATTAC-3′) and 541R (5′-CTCTGTGTTGACTGAATACAG-3′) with cycling conditions as previously described [Reference Allander2]. The amplicon size was 354 bp and the thermocycler used was ‘PTC-100’ (MJ Research Inc., Waltham, MA, USA). As positive control we used a HBoV PCR product cloned into the plasmid pCRII-TOPO XL (Invitrogen). The detection limit, determined by serial dilutions of that clone, was 40 copies of plasmid/reaction. HBoV amplicons were confirmed by sequencing of a conserved region (nucleotides 2281–2634) in the gene for the putative non-capsid protein 1 (NP-1). Sequencing reaction was performed using BigDye Terminator version 3.1 Cycle Sequencing (Applied Biosystems, Foster City, CA, USA) and the ABI Prism 377 DNA sequencer. DNA sequences were assembled and analysed with seqman, editseq, and megalign programs in Lasergene (DNASTAR, Madison, WI, USA).
All HBoV-positive NPA were further tested for other respiratory viruses. Reverse transcription–PCR (RT–PCR) was performed for HPIV 1, 2 and 3, Flu A and B, HCoV 229E and OC43, picornavirus and HMPV, and PCR was performed for HAdV. RT–PCR was performed using ImProm-II Reverse Transcriptase (Promega) according to the manufacturer's instructions. Random hexamers (Invitrogen) were used as RT primers, except for HCoV RT, where 229E-2 (5′-GACTATCAAACAGCATAGCAGC-3′) and OC43-2 (5′-GCAAAGATGGGGAACTGTGG-3′) specific primers [Reference Pitkäranta4] were used.
PCR for HPIV 1, 2 and 3 was performed in a multiplex format using Taq DNA polymerase (Biotools) primers Para1 (5′-CCTTAAATTCAGATATGT-3′), Pr1 (5′-GATAAATAATTATTGATACG-3′), Para2 (5′-AACAATCTGCTGCAGCAT-3′), Pr2 (5′-ATGTCAGACAATGGGCAAAT-3′), Para 3 (5′-CTGTAAACTCAGACTTGG-3′) and Pr3 (5′-TTTAAGCCCTTGTCAACAAC-3′) with cycling conditions as previously described [Reference Echevarría5]. Primers were designed for the haemagglutinin protein gene and amplicon sizes were 478 bp for HPIV 1 and 3, and 508 bp for HPIV 2.
PCR for Flu A and B was performed in a duplex format using Taq DNA polymerase (Biotools) primers AM149 (5′-CTCATGGAATGGCTAAAGACA-3′), AM501R (5′-TGCTGGGAGTCAGCAATCTG-3′), BM26 (5′-TGTCGCTGTTTGGAGACACA-3′), BM470R (5′-TGTGATGCTTGTTTTTCGCA-3′) with cycling conditions as previously described [Reference Ruest6]. Primers were designed for the matrix protein gene and amplicon sizes were 353 bp for Flu A and 469 bp for Flu B.
PCR for HCoV 229E and OC43 was performed using Taq DNA polymerase (Biotools) primers 229E-1 (5′-GGTACTCCTAAGCCTTCTCG-3′) and OC43-1 (5′-AGGAAGGTCTGCTCCTAATTC-3′) with cycling conditions as previously described [Reference Pitkäranta4]. Primers were designed for the nucleocapsid protein gene and amplicon sizes were 295 bp for 229E and 369 bp for OC43.
PCR for HMPV was performed using Taq DNA polymerase (Biotools) primers FF1 (5′-GAGCAAATTGAAAATCCCAGACA-3′) and FR1 (5′-GAAAACTGCCGCACAACATTTAG-3′) with cycling conditions as previously described [Reference Falsey7]. Primers were designed for the fusion protein gene and amplicon size was 347 bp.
PCR for AdV was performed using Taq DNA polymerase (Biotools) primers AV1 (5′-GCCGAGAAGGGCGTGCGCAGGTA-3′) and AV2 (5′-TACGCCAACTCCGCCCACGCGCT-3′) with cycling conditions as previously described [Reference Hierholzer8]. Primers were designed for the hexon gene and amplicon size was 162 bp.
For HRV detection, a PCR for picornavirus was performed using Taq DNA polymerase (Biotools) primers OL26 (5′-CGGACACCCAAAGTAG-3′) and OL27 (5′-Biot-CGGACACCCAAAGTAC-3′) with cycling conditions as previously described [Reference Pitkäranta4]. Primers were designed for the 5′ untranslated region gene and amplicon size was 392 bp. The picornavirus-positive PCR products were further tested in a hybridization assay, following previously published procedures and an enterovirus-specific probe (5′-GGCCGCCAACGCAGCC-3′) [Reference Pitkäranta4]. Samples positive for picornavirus and negative for EV by this assay were sequenced to confirm positivity for HRV.
HRSV RT–PCR was performed using ImProm-II Reverse Transcriptase (Promega), primed with random hexamers (Invitrogen) according to the manufacturer's instructions. PCR was performed in a semi-nested format using Taq DNA polymerase (Biotools) primed with FV (5′-GTTATGACACTGGTATACCAACC-3′) and GAB (5′-YCAYTTTGAAGTGTTCAACTT-3′) in the first round, and F1AB (5′-CAACTCCATTGTTATTTGCC-3′) and GAB primers in the second round. The PCR cycling conditions in both rounds were: 35 cycles of 94°C (1 min), 55°C (1 min), 72°C (1 min), with a final extension step of 10 min at 72°C. The primer FV [Reference Zheng9] was designed for the attachment G protein gene, while GAB [Reference Peret10] and F1AB [Reference Peret11] were for the fusion protein gene. Amplicon sizes were 489 bp for RSV-A and 492 bp for RSV-B.
Negative and positive controls were included in all PCR assays and PCR for β-actin was used as internal control for nucleic acid extraction in all tests. After amplification, PCR products were visualized on 1·5% agarose gels after staining with ethidium bromide and using the MultiImageTM Light Cabinet (Alpha Innotech Corporation, San Leandro, CA, USA). PCR preparations and product analysis were performed in separate rooms, with segregated materials and instruments.
A total of 262 NPA from 248 patients were collected in 2005. The male:female ratio was 1·1 (128 boys) and median age was 4 months (14 days to 50 months). HBoV was detected in 26 children (10·5%) and occurred year-round with peak activity in April and early autumn (Fig. 1). The male:female ratio of HBoV-positive children was 1·9 (17 boys), the median age was 8 months (1–36 months) and 88% were aged <2 years (58% <1 year).
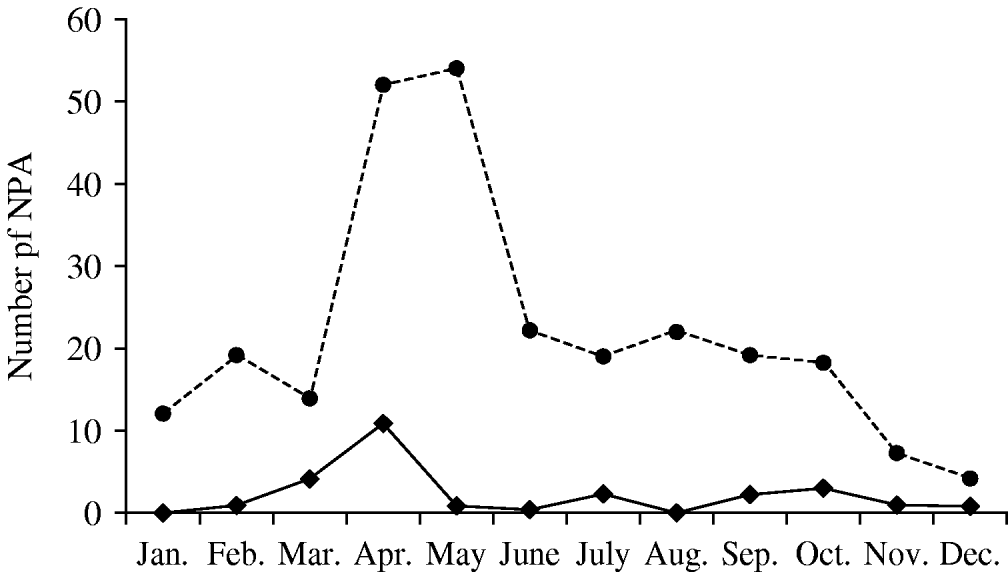
Fig. 1. Seasonality of human bocavirus (HBoV) infection in Ribeirão Preto, Brazil (2005). ◆––◆, HBoV-positive nasopharyngeal aspirates (NPA); •- - -•, all NPA tested.
Clinical data were available for 25/26 HBoV-positive children and indicated cough (92%) and dyspnoea (64%) as the most frequent findings, with coryza (36%), stuffy nose (28%), sneezing (16%) and apnoea (4%) being less frequent. Of 24 HBoV-positive children who were hospitalized, 13 (54%) received oxygen, two (8%) received mechanical ventilation and the period of hospitalization for length of stay was 7·5 (2–120) days. Final diagnoses of the 26 HBoV-positive children were: wheezing/asthma (46%), bronchiolitis (31%), non-specific ARI (11%), pneumonia (4%), whooping cough (4%) and bronchiolitis obliterans (4%). The clinical parameters observed for HBoV-positive children were not significantly different from the HBoV negative children.
Co-infections by other respiratory viruses were found in 21 (81%) of 26 HBoV-positive patients (Table 1). In the subset of five children who were positive only for HBoV, their predominant symptoms were cough (100%) and dyspnoea (80%) and these cases occurred throughout the year. In addition, wheezing was noted in two patients (40%) and coryza, stuffy nose and fever were each noted in one (20%) patient. Sneezing was not observed in these patients, and in two (40%) oxygen supplementation was needed. Their average hospital length of stay was 8 (5–31) days and their discharge diagnoses were wheezing in four and bronchiolitis in one patient.
Table 1. Respiratory viruses detected in HBoV-positive samples
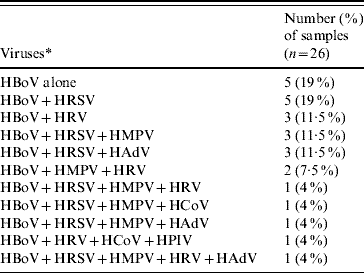
* For virus definitions see main text.
Nucleotide sequence analysis of the 26 HBoV amplicons (nucleotides 2281–2634 of the NP-1 gene) revealed high degree of identity (99·7%) with the prototype Swedish strains (ST1 and ST2, GenBank accession nos. DQ000495-DQ000496) [Reference Allander2].
We found HBoV DNA in 26 (10·5%) of 248 children aged <5 years admitted in 2005 with ARI in two hospitals in Ribeirão Preto, southeast Brazil. This frequency is similar to studies conducted in South Korea (10·3%) [Reference Choi12] and Germany (10·3%) [Reference Weissbrich13]. Although HBoV was detected year-round, a seasonal increase in circulation was observed in early autumn (Fig. 1).
A high rate (81%) of respiratory virus co-infection was found by RT–PCR in HBoV-positive patients, but this finding should be interpreted with caution. As previously shown for another respiratory virus, i.e. HRV, the viral RNA can be detected by RT–PCR in 17% of asymptomatic children [Reference Camara14]. This could have been due to the detection of viral genomes prior to the onset of symptoms or, more likely, to prolonged shedding after the symptomatic period. Nevertheless, the remarkably high rate (81%) of detection of other respiratory viruses along with HBoV highlights the need to consider viral co-infections in studies concerning the pathogenesis of individual viral respiratory infections.
The small number of patients with infection caused only by HBoV does not allow for definitive conclusions to be drawn about the clinical impact of this agent. There were no significant differences in the frequencies of either individual symptoms or final diagnosis between HBoV patients with and without co-infections. Of note, in the HBoV-only group, symptoms of the lower respiratory tract, mainly wheezing, were more frequent than cold-like, upper respiratory tract symptoms.
The NP-1 gene nucleotide sequence showed a high degree of homology in the samples in the present study and the prototype Swedish ones [Reference Allander2]. However, it must be noted that the sequence analysis was performed in a region generally considered to be conserved, as opposed to the VP1 region, which seems to contain most of the variability and has provided the basis for the establishment of two HBoV lineages.
ACKNOWLEDGEMENTS
This study was supported by the São Paulo State Research Foundation (FAPESP) through the Viral Genetic Diversity Network Programme (VGDN) and by the Brazilian National Research Council (CNPq).
DECLARATION OF INTEREST
None.




