INTRODUCTION
Measles is a highly infectious disease that can result in serious complications. The only way to prevent infection is through measles vaccination. In Greece, the MMR vaccine was introduced in the National Immunization Programme in 1989 and the overall incidence of measles has since fallen. A total of 12 251 measles cases were reported between 1990 and 2000, 636 cases were reported during the 2005–2006 outbreak and 126 cases reported during the current outbreak (2010). The vaccination scheme was reconsidered and has been changed twice since 1999, as two rubella outbreaks in 1993 and 1999 and a measles outbreak in 1995–1996 occurred. The current scheme consists of two doses of the vaccine, at ages 12–15 months and 4–6 years [Reference Farmakis1]. The initial European goal for measles virus (MV) elimination by 2007 was reset to 2010 due to continuous outbreaks reported from different European countries since 1998 [2–5]. The new goal for MV elimination is now set to 2015.
Greece has experienced measles outbreaks in 2006 and 2010. Molecular analysis of the 2006 measles outbreak showed that measles D6 genotype strains were responsible for the outbreak in northern Greece, while strains of the D6 and D4 genotypes were co-circulating in the south [Reference Gioula6, Reference Kokotas7]. The current measles outbreak has been ongoing since January 2010, with a total of 126 cases, reported up to July 2010 to the Hellenic Center for Disease Control and Prevention (HCDCP) through the mandatory notification system. Patients were mostly unvaccinated and belonged to three groups: Roma population of Bulgarian nationality, Greek Roma population and Greek non-minority population. In these population groups, 67% (n=24), 95% (n=41) and 25% (n=11) of cases, respectively, were children aged 0–14 years [Reference Pervanidou8]. European countries with the goal of measles elimination by 2015 are expected to reach <1 confirmed measles case per million population per year. To reach this goal, they are expected to achieve MV vaccination of 95% for the first dose and at least 80% for the second dose [5, Reference Kremer9]. The incidence of measles infection in the general Greek population is so far 1·1 cases/100 000 individuals. Another criterion for MV elimination in Europe, which has been achieved in Greece since 2000, is that the length of transmission chains is <12 months. However, as the year targeted for MV elimination was reached a new outbreak occurred in Greece; a revised target has now been set for 2015. The purpose of this study was the molecular and phylogenetic analysis of the Greek MV circulating strains.
METHODS
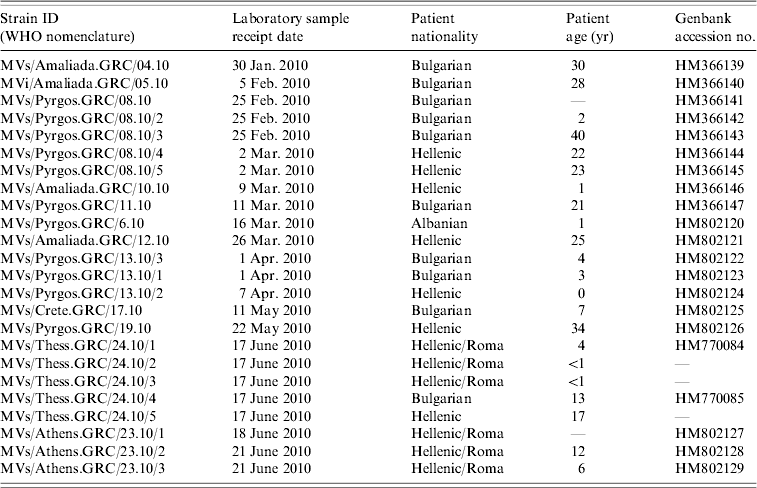
The examined samples were 24 nasopharyngeal swabs that were collected at southern and northern Greek hospitals during the period January–July 2010. Serum from the same patients tested positive for IgM antibodies (Table 1). Samples were initially examined for the presence of MV RNA. Molecular virus detection and further molecular analysis were performed at the National Measles/Rubella Laboratory at the Hellenic Pasteur Institute and at the 2nd Laboratory of Microbiology, Medical School, Aristotle University of Thessaloniki.
Table 1. Epidemiological information of clinical samples
Reverse transcriptase–polymerase chain reaction (RT–PCR)
Viral RNA was extracted from the clinical samples using the QIAamp viral RNA mini kit (Qiagen, Germany). RNA samples were converted into cDNA, and amplified using the Superscript one step RT–PCR kit (Invitrogen, UK) [10]. Specifically, primers PARMV60 and PARMV63-3 (Invitrogen) were used to amplify the 600-bp fragment of the 3′-end terminus of the nucleoprotein gene of the virus, as described in the CDC protocol [10].
PCR product purification and nucleotide sequence determination
PCR products were separated with gel electrophoresis and individual bands were purified with the gel extraction kit (Invitrogen). Primers PARMV60F and PARMV63-3R10 were used for sequencing with a GenomeLabDTCS-Quick Start sequencing kit, on a genetic analyser CEQ™ 8000, Beckman Coulter. All the strains were genotyped by sequencing at least 450 nt that code for the 3′-end terminus of the N gene hypervariable region of the MV genome in forward and reverse direction, as recommended by WHO [11].
Nucleotide distances and phylogenetic analysis
The obtained Greek N-gene sequences were aligned with those of WHO reference strains, using ClustalW software [Reference Larkin12]. Phylogenetic relationships were inferred from the aligned nucleotide sequences by applying the MEGA 4.1 program [Reference Tamura13]. Greek viral N-gene sequences were also compared in terms of their mean sequence diversity, by pairwise alignments of the Greek isolates and representative WHO reference strains [Reference Kimura14]. The obtained sequences were also analysed in terms of closest homologue sequence using BLAST software (NCBI).
Nucleotide sequence accession numbers
The active reference strains used in this study are those proposed by WHO [15]. Representative sequences have been deposited in the GenBank/EMBL/DDBJ databases under accession numbers: HM770084-85, HM366139-147 and HM802120-26.
RESULTS
Nucleotide distance analysis and phylogeny
All viruses detected in Greece clustered with the D4 reference strain (Fig. 1) Subgroups were also observed in the Greek viruses and were further analysed. Specifically, MV was isolated from 10 patients of Bulgarian origin and 90% of those isolated strains were identical to each other, with 2·6% nucleotide difference from the reference strain Montreal.CAN/89. Only one isolate of Bulgarian origin, MV/Amaliada.GRC/04.10 differed by 1 nt (0·22%) from the rest. Moreover, six isolates that derived from the Roma community, both from the north and from the south of Greece, varied from those that derived from individuals of Bulgarian nationality. Specifically, three of the isolates were northern Greek strains (MVs/Thess.GRC/24.10/1, MVs/Thess.GRC/24.10/2, MVs/Thess.GRC/24.10/3), with 4 nt variation and genetic distance 3·5% from Montreal.CAN/89 that were not previously identified as revealed by BLAST search. The other Roma-derived strain was from southern Greece (MVs/Athens.GRC/23.10), and had 2 nt variation from the Bulgarian-origin Greek isolates and 2·85% genetic distance from the reference strain. The remaining two southern Greek strains (MVs/Athens.GRC/23.10/2, MVs/Athens.GRC/23.10/3), which also belonged to the Roma community, differed by 1 nt from the rest of the Bulgarian-origin Greek strains and had 2·85% genetic distance from Montreal.CAN/89. There was also one strain isolated from a patient of Albanian nationality (MVs/Pyrgos.GRC/6.10), which differed by 1 nt from the rest of the Hellenic or Bulgarian strains that were isolated from the same region. Last, the MV isolates from patients that belonged to the southern and northern Greek non-minority population, and in particular MVs/Pyrgos.GRC/08.10, MVs/Amaliada.GRC/10.10, MVs/Amaliada.GRC/12.10, MVs/Pyrgos.GRC/13.10/2, MVs/Pyrgos.GRC/19.10 and MVs/Thess.GRC/24.10/5 were all identical with nucleotide divergence of 2·67% (12 nt) from Montreal.CAN/89 and only 0·22% nucleotide divergenece (1 nt) from the Albanian isolate MVs/Pyrgos.GRC/6.10. The maximum nucleotide divergence between the sequences of the 24 studied Greek D4 strains was 0·88% (4 nt).
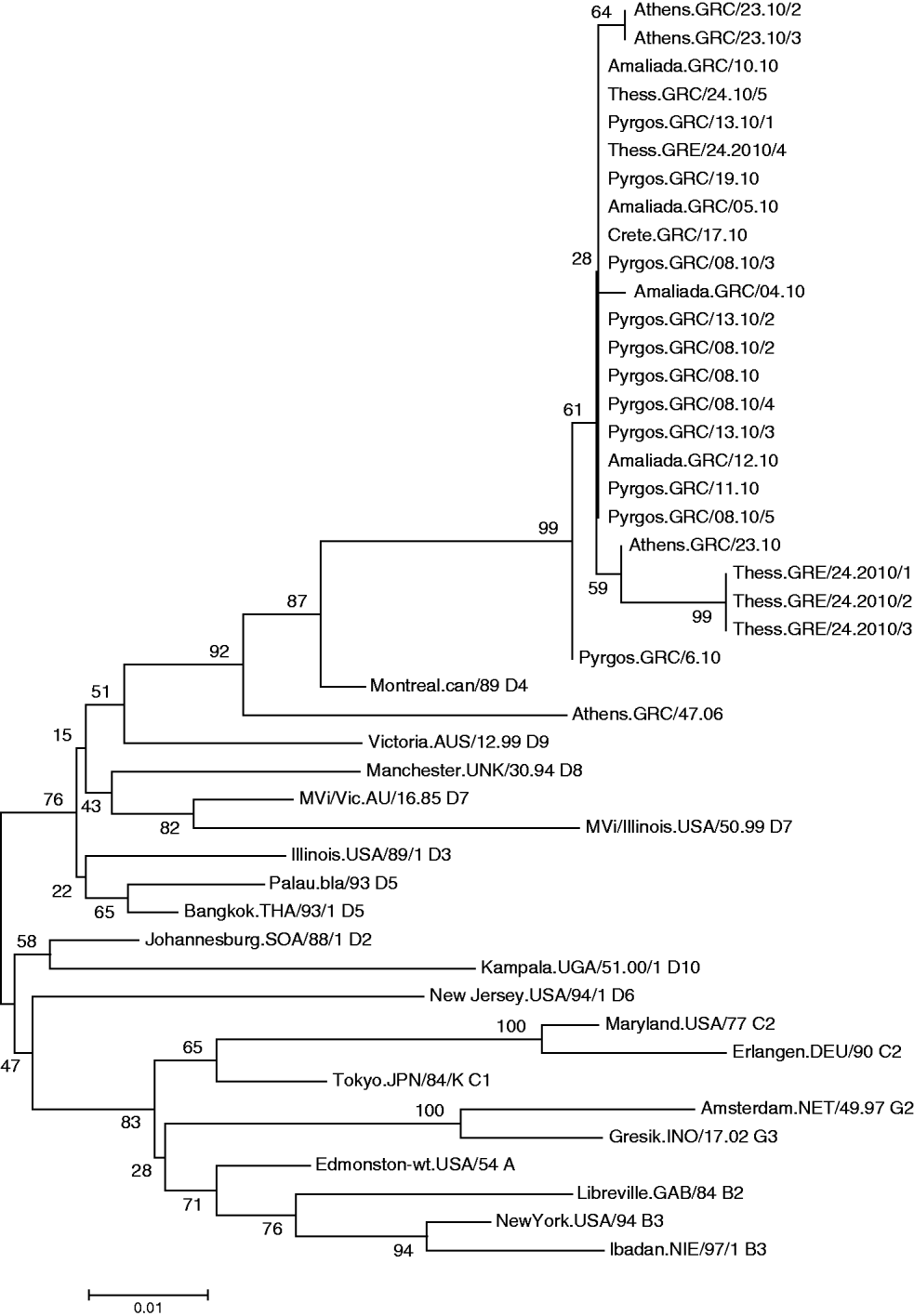
Fig. 1. Phylogenetic analysis of the sequences of the Greek measles strains based on the 450 carboxy-terminal end of the N gene was achieved by MEGA 4.1 software. The dendrogram was constructed by comparison of the Greek D4 sequences with reference strains [11]. The tree was built with the neighbour-joining algorithm and evaluated by 1000 bootstrap pseudoreplicates. All viruses detected clustered with the D4 reference strain.
Moreover, comparing the four suggested subclusters of D4 circulating strains [Reference Kremer9], with the Greek D4 genotyped strains, it was observed that the D4 MV responsible for the outbreak in Greece is more closely related to the isolates from subgroup 4 (Fig. 2). Isolates from Greece possibly share a common evolutionary history with other clinical strains isolated around Europe during this epidemic period. Strains isolated in the UK, Ireland, Spain, Skopje, Switzerland and Turkey cluster with the subgroup 4 strains isolated in Greece (Fig. 2).
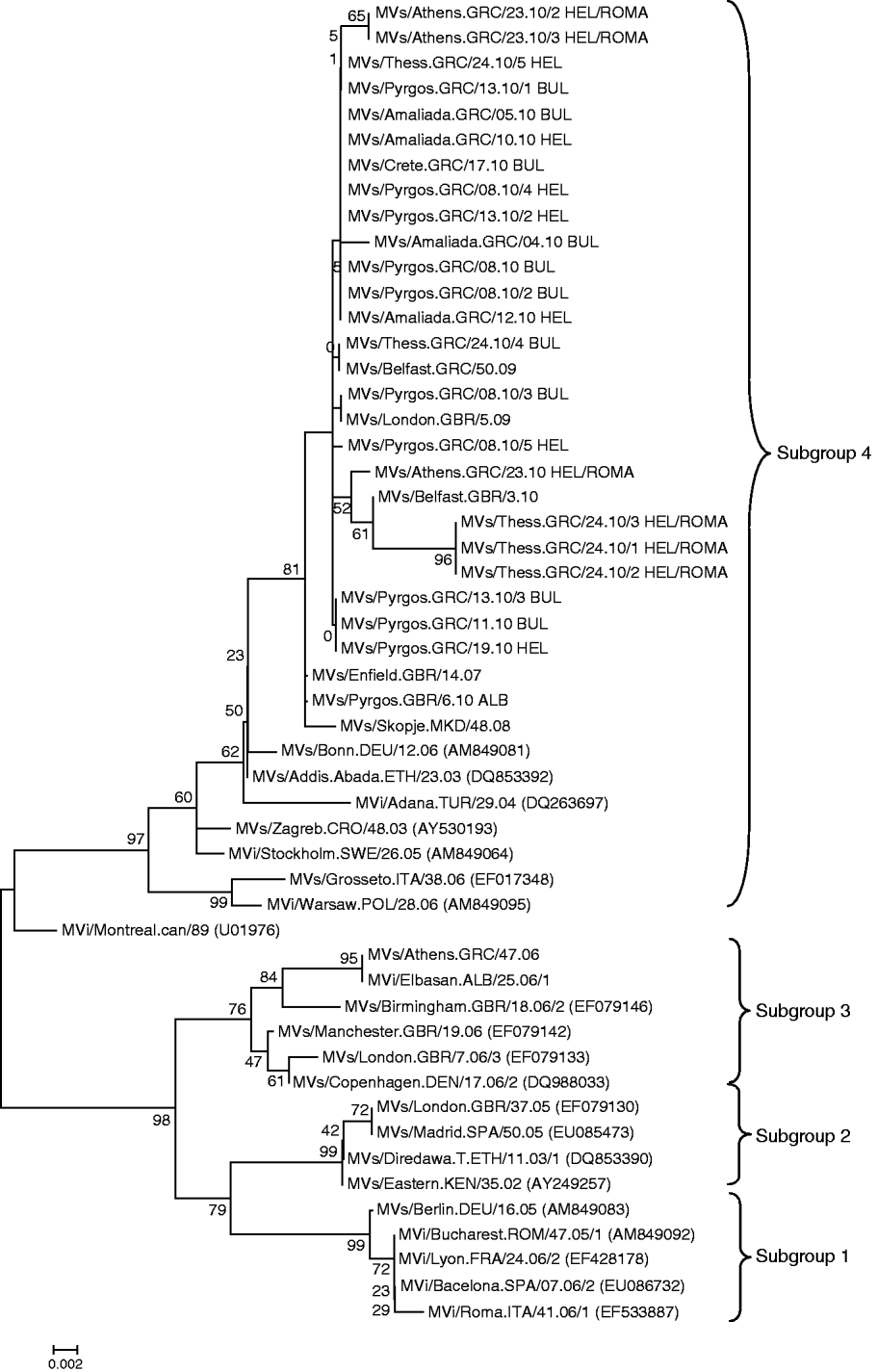
Fig. 2. Phylogenetic analysis of the sequences of the Greek measles strains based on the 450 carboxy-terminal end of the N gene was achieved by MEGA 4.1 software. The dendrogram was constructed by comparison of the Greek D4 sequences with representative strains from the four proposed subgroups of the D4 genotype and currently circulating D4 strains from other countries [Reference Kremer9]. The tree was built with the neighbour-joining algorithm and evaluated by 1000 bootstrap pseudoreplicates. All Greek 2010 isolates belong to subgroup 4.
DISCUSSION
In the present report, the nucleotide and phylogenetic analysis of MV detected in Greece revealed genotype D4. It of note that the D4 cluster was also found in the southern part of Greece during the previous measles epidemic, in 2005–2006, and included 10 strains all identical to each other and with 3·8% genetic distance from Montreal.CAN/89, which is the WHO D4 reference strain [Reference Kokotas7]. Interestingly, the Greek 2005–2006 D4 strain showed a 5·5–6·4% genetic distance from the present circulating D4 strains, which indicates the possibilty that D4 circulation was interrupted from 2006 to 2009 and a new transmission chain initiated. However, most of the Greek measles strains during the 2005–2006 outbreak were of D6 genotype [Reference Gioula6, Reference Kokotas7], which has also disappeared probably due to the enhanced vaccination strategy that followed.
It appears that the 2010 Greek D4 isolates form subgroups, depending upon their origin. As reported by HCDCP, the 126 reported cases belonged to one of the three groups: the Roma population of Bulgarian nationality, the Greek Roma population and the Greek non-minority population [Reference Pervanidou8]. Most cases belonged to one of the two minority groups and a possible explanation for this phenomenon might be the low vaccination coverage of people with increased mobility, which can lead to the maintenance of circulation of the virus and its transmission. In fact, according to several studies carried out in 2003–2005 in different parts of Greece, vaccination coverage of preschool children, schoolchildren, and adolescents with one dose of MMR vaccine is >95% in the non-minority population and 80–90% in children of immigrant families. Vaccination with two doses covers about 60–80% of the general child population, but vaccination coverage in Roma families has been found to be very low (2–12%) despite the number of vaccination campaigns that took place in this group during the previous years [Reference Gioula6].
Concerning the D4 strains that caused the 2010 outbreak in Greece, increased differentiation of the Greek D4 variant from the prototype D4 Montreal.CAN/89 strain was observed. The percentage of the maximum nucleotide divergence was estimated at 3·5%. In a more detailed phylogenetic analysis of the Greek D4 strains in relation to the four subgroups that have been proposed by WHO and that are circulating worldwide, the 2010 Greek D4 variants clustered with subgroup 4 variants [Reference Kremer9, Reference Mbugua16], whereas the 2006 Greek D4 variants clustered with subgroup 3 stains [Reference Kremer9, Reference Mbugua16] (Fig. 2). Interestingly, isolates from the Greek non-minority population were almost identical, with only 1 nt difference from the Albanian isolate. Additionally, the percentage of maximum nucleotide divergence between the 2010 Greek variants was 0·88%. This nucleotide variation showed that it is possible that isolates, coming from north or south, Roma or the general population could belong to the same transmission chain, with circulation that was observed for 6 months. However, because of the presence of nucleotide diversity between MV isolates from Bulgarian and the Greek Roma populations, we cannot exclude the possibility that two transmission chains existed. However, since there were no deposited sequences identical to the most divergent Greek isolates, derived from individuals of the Roma community of Hellenic nationality, this observation cannot be confirmed. The accurate identification of the Greek D4 measles strains' source and the exact route was also not feasible, due to the lack of important sequence information from public databases. There is nevertheless a clear epidemiological link of the Greek outbreak to the recent Bulgarian outbreak, which started in April 2009 [Reference Pervanidou8, Reference Marinova, Kojouharova and Mihneva17].
On the other hand, genotype D4 viruses are still endemic on the Indian subcontinent as well as in East and South Africa [Reference Mulders, Truong and Muller18–Reference Muller20]. Genotype D4 has been repeatedly identified in the WHO Eastern Mediterranean Region [Reference Djebbi21] as well as in outbreaks and sporadic cases in several European countries. Germany reported an outbreak early in 2009. Epidemiological analysis has revealed that a strain transported from Hamburg spread the MV to Bulgaria [Reference Marinova, Kojouharova and Mihneva17]. Other European countries, have reported similar outbreaks since then [Reference Kasper22–24]. The Greek outbreak is epidemiologically linked to the Bulgarian one. In all cases the Roma community seems to have played a significant role in virus transportation, due to the high mobility and the low vaccination coverage of this population [Reference Lopalco and Martin4]. Euvac.net reported a crude incidence of 2·05 confirmed MV cases/100 000 inhabitants, which is still far from the elimination goal of <1 confirmed case/1 000 000 inhabitants [Reference Lopalco and Martin4].
As already mentioned, most of the Greek isolates were epidemiologically linked to the extended Bulgarian measles outbreak, and half of the affected individuals had a history of travel to Bulgaria or Bulgarian nationality [Reference Marinova, Kojouharova and Mihneva17]. Furthermore, some Greek strains showed an exact nucleotide match with London.GBR/5.09 and Belfast.GBR/50.09 isolates and 1% nt variation from other 2009 isolates from Italy, France and the UK, and from 2008 isolates from Spain. Regarding, the other MV subgroup that was detected in northern Greece, which seem to originate from the Greek Roma community, no previously identified exact sequence match could be identified during the BLAST search. The closest homologous strain was detected in Ireland in 2010, and had 1% N-gene sequence variation. In previous years this strain circulated in the UK and other European countries.
It should be noted that paediatricians in Greece have observed a 40% decrease in MMR infant and child vaccinations this year (media reports). This has been attributed to doubts regarding the safety of vaccines that pandemic influenza vaccination has spread to the public. This is likely to contribute to a more widespread circulation, rather than a localized limited transmission of the virus in Greece in the future.
The importance of the genetic characterization of the circulating MV strains is emphasized, as it helps to distinguish between continuous circulation from importation and limited transmission of the viruses in a certain region [Reference Mulders, Truong and Muller18, Reference Rota19]. Knowledge of the genotypes that are currently circulating in Greece and in neighbouring countries will aid future studies of measles transmission and monitor the success of measles vaccination programmes in Greece, which will eventually lead to the elimination of MV.
DECLARATION OF INTEREST
None.





