INTRODUCTION
Norovirus (NoV) is the leading cause of non-bacterial, acute gastroenteritis and has been associated with gastroenteritis in infants, young children and adults in developed as well as developing countries [Reference Fankhauser1–Reference Glass, Parashar and Estes4]. NoVs are non-enveloped, icosahedral viruses and possess a linear, positive-sense, single-stranded RNA (ssRNA) genome of about 7·4–7·7 kb. NoV belongs to the Caliciviridae family. The RNA genome of NoV is organized into three major open reading frames (ORF1, -2, -3) with a polyadenylated 3′-end [Reference Jiang5]. Furthermore, ORF1 encodes a large polyprotein that is proteolytically processed into the mature non-structural proteins [Reference Liu, Clarke and Lambden6]. ORF2 encodes the major capsid protein, VP1, and ORF3 encodes a minor structural protein, VP2. NoV has five major phylogenetic clades, or genogroups, designated GI to GV [Reference Zheng7]. The prevalence of antibody to the GII viruses (Mexico, Hawaii, or Lordsdale) appears to be greater than that of the GI viruses in most studies [Reference Glass, Parashar and Estes4, Reference Cubitt, Green and Payment8]. Like all enteric viruses, NoVs are associated with several modes of transmission. The predominant modes of transmission are person-to-person contact and food- or water-borne transmission [Reference Fankhauser1, Reference Boxman9]. Clinical manifestations associated with NoV infections include nausea, vomiting, diarrhoea, abdominal cramps, headache, fever (subjective), chills, myalgia and sore throat. Vomiting was more frequently associated with diarrhoea in children, whereas in adults the reverse was observed. The duration of illness may range from 2 h to several days, with a mean or median of between 12 h and 60 h [Reference Kaplan10]. RT–PCR, coupled with sequence analysis of amplicons, has been used extensively to detect and characterize NoVs in various outbreaks. A preliminary report in 2000 showed that NoVs were associated with diarrhoea in the coastal region of southern India [Reference Kang11]. Since then many studies from different parts of the country showed NoV as another important cause of gastroenteritis. This study reports the detection and molecular characterization of NoVs, which play an important role as a viral aetiological agent of acute watery diarrhoea besides rotavirus in eastern India. The objectives of the study were (1) to describe the epidemiological behaviour of NoVs in Kolkata during a 2-year study including seasonal and clinical patterns, (2) to show co-infections with other pathogens such as bacteria, parasites and other viruses, namely rotaviruses, adenovirus, astrovirus or sapovirus, and (3) to understand the phylogenetic relationship of NoV detected in Kolkata with other NoV strains reported from other countries.
MATERIALS AND METHODS
Study design
This hospital-based study was performed by collecting stool specimens from patients admitted to Infectious Diseases & Beliaghata General (ID&BG) Hospital, Kolkata, India, with acute gastroenteritis from November 2007 to October 2009. The ID&BG Hospital provides treatment to about 20 000–25 000 acute diarrhoea patients annually. Patients admitted to the hospital with diarrhoeal complaints were included in the study using a systematic sampling process: on two randomly selected days each week every fifth patient with diarrhoea or dysentery was enrolled. The system remained unbiased for sex and age of the patient at the time of selection. A faecal sample from each of the enrolled patients was collected, processed for the study and total of 2495 faecal samples were analysed during the study period. After informed consent was given epidemiological information from each case was obtained from patients or guardians. Clinical data involving such disease manifestations as fever, vomiting, abdominal pain, or bloody diarrhoea were collected for all patients. Severity criteria, such as duration of diarrhoea, number of stools or vomiting episodes, range of body temperature, and degree of dehydration, were determined for all enrolled patients. This study was conducted with prior approval of the ethics committee of National Institute of Cholera and Enteric Diseases, Kolkata.
Sampling and preliminary analysis
Faecal specimens were collected in sterile glass containers and transported at 4°C to the Division of Virology, National Institute of Cholera and Enteric Diseases, Kolkata. All faecal specimens were collected within <24 h after admission. In a 2-ml sterile Eppendorf tube 30% faecal suspension was prepared with 1× PBS (pH 7·2). The tubes containing faecal suspension were stored at 4°C for further processing. All samples were simultaneously screened for Vibrio cholerae O1, V. parahaemolyticus, V. fluvialis, Escherichia coli, Shigella spp., Aeromonas spp., Campylobacter coli and Salmonella spp. by conventional bacterial culture procedures, and diarrhoeagenic E. coli (DEC) types [enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC), and enterohaemorrhagic E. coli (EHEC)] were studied by PCR-based methods. Parasites (Cryptosporidium, Entamoeba histolytica, Girardia lamblia) were screened by using conventional microscopy and staining methods.
RNA extraction
Extraction of viral RNA was performed using QIAamp viral RNA mini kit (Qiagen, Germany) according to the manufacturer's instructions. Sixty microlitres of molecular biology grade viral RNA was eluted for use in RT–PCR experiments. The viral RNA was stored at −20°C (for immediate use) or at −80°C for long-term storage.
Reverse transcription
Briefly, 10 μl RNA was placed in a 0·2-ml Eppendorf tube and 1 μl of random primer (150 ng/μl; Invitrogen, USA) added. The Eppendorf tube containing the RNA and random primer was incubated in a thermal cycler at 65°C for 5 min and then kept on ice to snap chill for 10 min, followed by addition of 9 μl RT mix to adjust the final volume to 20 μl. RT mix was comprised of 4 μl of 10× reverse transcriptase buffer (New England BioLabs, USA), 1 μl of 0·1 m DTT (dithiothreitol), 2·5 μl of 10 mm dNTPs (New England BioLabs), 1 μl RNase inhibitor (40 U/μl, Ambion, USA), 0·5 μl Moloney murine leukaemia virus reverse transcriptase (MMLV, 200 U/μl, New England BioLabs). The RT reaction was performed for 60 min at 42°C to produce cDNA; an aliquot was used directly in the PCR amplification; excess was stored at −20°C or −80°C for immediate or later use, respectively.
Region C PCR amplification
Five microlitres of cDNA was added to the PCR mix containing 2·5 μl of 10× PCR buffer (Invitrogen, USA), 0·75 μl of 50 mm MgCl2 (Invitrogen), 0·5 μl of 10 mm dNTPs (New England BioLabs), 1 μl of 10 pmol of each GI-specific primer (G1SKF and G1SKR) or GII primer (G2SKF and G2SKR) [Reference Kojima12], 0·25 μl Taq DNA polymerase (5 U/μl, Invitrogen) and 14 μl RNase-free water to a final volume of 25 μl. PCRs in separate tubes for NVGI and NVGII were performed under the following conditions: 94°C for 3 min followed by 40 cycles of 94°C for 30 s, 46°C for 30 s, and 72°C for 1 min, with a final extension step at 72°C for 7 min. The 330-bp and 344-bp amplicons obtained from NVGI and NVGII viruses, respectively, were visualized by 2% agarose gel electrophoresis followed by ethidium bromide staining.
PCR product purification
Twenty-four NoV GII-positive samples showing expected amplicon size (GII: 344 bp) were subjected to 50 μl PCR reaction and the final PCR product was purified using QIAquick PCR Purification kit (Qiagen). The PCR purified product was then used for sequencing.
Nucleotide sequencing
The purified PCR products were subjected to cycle sequencing using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit, version 3.1 (Applied Biosystems, USA). Sequencing was performed for region C in order to identify the exact genogroup as well as to identify recombinant viruses. For region C, sequencing was carried out by using both forward and reverse primers separately. The sequences were collected from an automated DNA sequencer (ABI 3100, Applied Biosystems). The nucleotide sequences reported in this study were deposited at the DNA Data Bank of Japan (DDBJ).
Phylogenetic analysis
Sequence identity was determined through BLAST (www.ncbi.nlm.nih.gov/blast) and multiple sequence alignment was carried out with the CLUSTAL W program [Reference Thompson, Higgins and Gibson13]. The phylogenetic analysis of aligned sequences was carried out using MEGA 4.0 [Reference Tamura14]. The phylogenetic tree was generated with neighbour-joining algorithm [Reference Saitou and Nei15]. The reliability of the phylogenetic tree was tested by applying the bootstrap test with 1000 bootstrap replications.
Nucleotide sequence accession number
The partial nucleotide sequences of the gene encoding the capsid region of the NoV strains from Kolkata (present study) were submitted to DDBJ, under accession numbers AB539140–AB539163.
Statistical analysis
The data from the above identification procedures and the epidemiological survey were entered into a pre-designed pro-forma in the SQL server with a built-in entry validation checking facilitated program. Data were randomly matched and checked to establish consistency and validity. SPSS version 14.0 (SPSS Inc., USA) was used for statistical analysis.
RESULTS
During the period of study, NoVs (n=78, 3·12%) were detected by RT–PCR in diarrhoeic patients (n=2495, enrolled in the surveillance) at ID&BG Hospital. NoVs were sporadically detected throughout the year, and the number of positives detected was insufficient to determine seasonality (Fig. 1). Six (7·7%) out of 78 positive specimens belonged to NVGI and 72 (92·3%) specimens belonged to NVGII strains. NVGI infections were detected in November 2007 (1), February 2008 (3); March 2008 (1) and May 2009 (1), respectively. Of the diarrhoea cases, NoV was seen as either a single aetiological agent in 22/78 cases (28·2%) or found in association with other diarrhoeagenic pathogens, e.g. viral, bacteria, or parasitic agents in the other 56/78 cases (71·8%). The faecal specimens collected from all NoV-positive cases were cross-checked for any other co-infection with other enteric pathogens, such as diarrhoeagenic viruses, e.g. rotavirus, astrovirus, adenovirus, sapovirus or bacteria and parasites during the study.
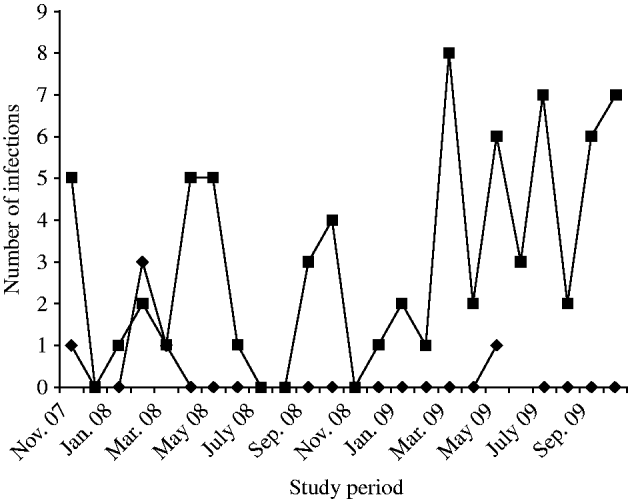
Fig. 1. Detection of NVGI (–◆–) and NVGII (–▪–) infections in diarrhoea cases in Kolkata, India from November 2007 to October 2009.
Of the 78 NoV-positive cases, at least one more pathogen was detected in 56 cases with rotavirus being the most common co-infection followed by Vibrio cholerae O1, Cryptosporidium spp. and E. coli (ETEC). The different combinations of mixed infection with different enteric virus, bacteria and parasites is shown in Table 1. Six instances of NoV infection showed co-infection with bacteria and parasites.
Table 1. Co-infection of norovirus with other enteric pathogens, i.e. viruses, bacteria and parasites in Kolkata, India during the study (November 2007 to October 2009)

* Rotavirus co-infection only with norovirus was seen in IDH 41, 186, 278, 434, 969, 2035, 2328, 2373, 2443, 2490.
Prevalence in different age groups and sex
Distribution of NoV gastroenteritis in children aged <1 and >1–2 years was similar, with the highest predominance of NoV in this age group. In the 14–30 years age group comprising older children and adults, the prevalence of NoV was marginally higher than that of children aged <2 years. Children aged between 2 and 14 years and adults aged between >30 years and >60 years showed lower rates of NoV positivity (Fig. 2). The difference in the NoV infection rate between males and females was significant (64·1% and 35·9%, respectively). Urban people of Kolkata showed more positivity for NoV infection than their rural counterparts (data not shown).
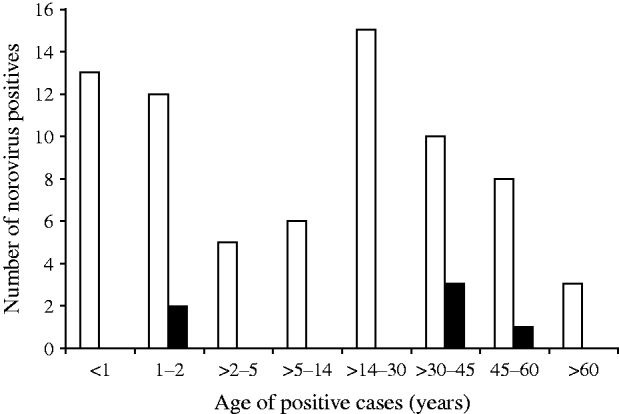
Fig. 2. Age distribution of norovirus positives detected in Kolkata, India from November 2007 to October 2009. ▪, NVGI; □, NVGII.
Comparison of clinical symptoms during NoV infections
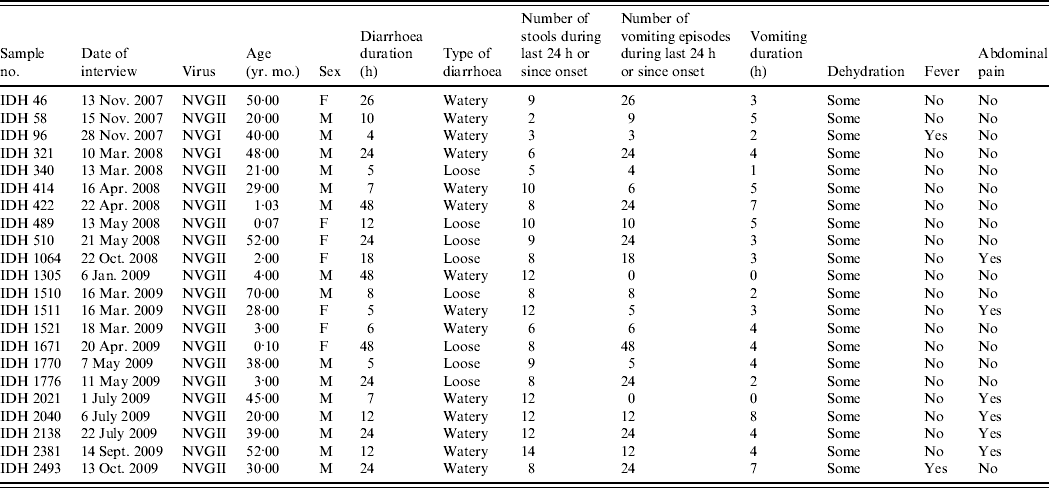
Clinical symptoms associated with diarrhoea such as number of stools, number of vomiting episodes and degree of dehydration were recorded in all diarrhoea cases, and relevant data for diarrhoea cases with NoV as the only detectable pathogen are shown in Table 2. All the patients had diarrhoea associated with some dehydration, two patients had fever and 16 (72·7%) patients showed no abdominal cramp or pain. Eighteen patients (81·8%) had >5 vomiting episodes in a 24-h period; two patients had no vomiting. Of 22 patients, seven (31·8%) were aged <5 years; interestingly the severity of NoV infection was greater in five (71·4%) of these cases who suffered >10 vomiting episodes and >8 stools during a 24-h period.
Table 2. The clinical details of patients showing sole norovirus infection in Kolkata, India during November 2007 to October 2009
Molecular characterization of partial capsid gene
It was observed that phylogenetic analysis of the 282-bp region of the capsid gene of the Kolkata strains reported in this study revealed they were divergent and occupied several branches of the phylogenetic tree (Fig. 3). It was observed that eight Kolkata strains clustered with Bristol/GII.4 (CAA54134) followed by the seven Kolkata strains with Fayetteville/GII.13 (AAM56034) and five Kolkata strains with Seacroft/GII.6 (CAB89101). Strain IDH340 clustered with Hawaii/GII.1 (AAB97768), IDH1521 clustered with Tiffin/GII.16 (AAS86789) and IDH495 and IDH500 clustered with Toronto24/GII.3 (AAA18930).
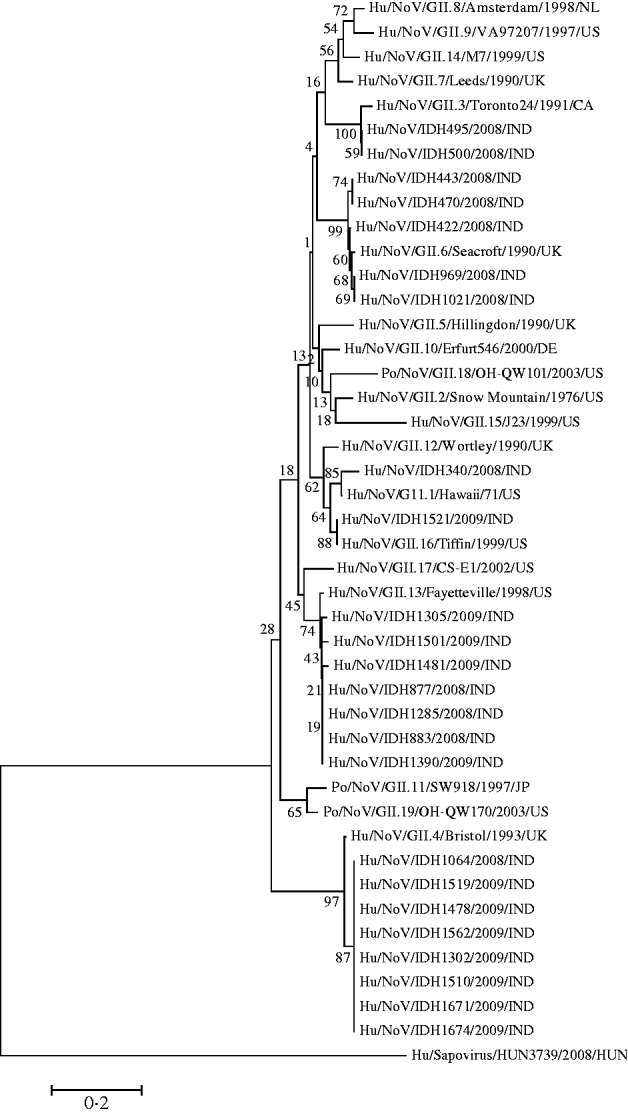
Fig. 3. Phylogenetic analysis based on deduced amino-acid sequences corresponding to 282-bp nucleotide fragment of the capsid gene of Kolkata NoV strains (shown in bold face) and other NoVs. The accession numbers of the capsid fragments for NoV strains shown on the tree is as follows: GII.8/Amsterdam (AAF05820); GII.9/VA97207 (AAK84676); GII.14/M7 (AAN05735); GII.7/Leeds (CAB89089); GII.3/Toronto24 (AAA18930); IDH495 (AB539144); IDH500 (AB539145); IDH443 (AB539142); IDH470 (AB539143); IDH422 (AB539141); GII.6/Seacroft (CAB89101); IDH969 (AB539148); IDH1021 (AB539149); GII.5/Hillingdon (CAB89088); GII.10/Erfurt546 (AAL18874) GII.18/OH-QW101 (AAX32877); GII.2/Snow Mountain (AAB16915); GII.15/J23 (AAN05736); GII.12/Wortley (CAB89099); IDH340 (AB539140); GII.1/Hawaii (AAB97768); IDH1521 (AB539160); GII.16/Tiffin (AAS86789); GII.17/CS-E1 (AAS86786); GII.13/Fayetteville (AAM56034); IDH1305 (AB539153): IDH1501 (AB539157); IDH1481 (AB539156); IDH877 (AB539146); IDH1285 (AB539151); IDH883 (AB539147); IDH1390 (AB539154); GII.11/SW918 (BAB83516); GII.19OH-QW170 (AAX32883); GII.4 Bristol (CAA54134); IDH1064 (AB539150); IDH1519 (AB539159); IDH1478 (AB539155); IDH1562 (AB539161); IDH1302 (AB539152); IDH1510 (AB539158); IDH1671 (AB539162); IDH1674 (AB539163); out group strain Sapovirus Kecskemet/HUN3739 (ACO72592).
DISCUSSION
The disease burden of NoVs has been well characterized for developed countries; however, there is little information about this virus in the Indian subcontinent. In this study, we report on the surveillance of NoV from 2495 cases of diarrhoea enrolled in the study following their hospitalization at ID&BG Hospital for treatment. In the present study, 78/2495 diarrhoea cases collected during the study period were found to be positive for NoVs by RT–PCR. In a study from Pune, India, it was reported that the detection of NoV in children aged <7 years was 10·7% [Reference Chhabra16]. In a study from Djibouti (Horn of Africa) researchers were able to detect eight NoV positives out of 75 cases in adults presenting with acute diarrhoea [Reference Maslin17]. In a 2-year study from outpatient clinics in Sapporo, Japan, Nakanishi et al. [Reference Nakanishi18] found 13·2% positivity for NoVs from 877 rectal swabs collected from patients and tested by RT–PCR; frequent vomiting was prominent in NoV gastroenteritis [Reference Nakanishi18]. Another study covering eight different cities of the Russian Federation described the results of 2-year surveillance for group A rotaviruses and other enteric agents, in patients hospitalized with acute gastroenteritis; whereas in the <5 years age group NoV positivity was 12·5%, while NoV was the pathogen most commonly detected in adults at a positivity rate of 11·9% [Reference Podkolzin19]. In 2008 a study from Tunisia showed 17·4% of NoV in acute infantile gastroenteritis cases in Tunisia [Reference Sdiri-Loulizi20]. To date, in India most of the studies on NoV gastroenteritis were performed on children. This study focuses on the NoV prevalence on both children and adults. Although reports from developed countries indicate that 5–12% of cases of sporadic gastroenteritis are associated with NoVs we observed 0·9% positivity (22/2945) for NoVs as the only detectable pathogen in this study. It is presumed that this may be related to immunity and the resultant mild or asymptomatic infection associated with many exposures to the virus through close contact with infected children, impurity of the water supply and environmental contamination. Of the 78 NoV-positive cases at least one more pathogen was detected in 56 cases with rotavirus being the most common co-infection followed by V. cholerae O1, Cryptosporidium spp. and E. coli (ETEC). In a study from Nicaragua, NoV was detected in 12% (65/542) of children; of these an important proportion (57%) of NoV-infected children were co-infected with diarrhoeagenic E. coli [Reference Bucardo21]. In our study we too found an important proportion of diarrhoeagenic E. coli co-infection with the NoV-positive cases. In a Korean study of children with acute diarrhoea it was reported that 33 (58·9%) NoV-positive cases showed mono-infection and 23 (41·1%) had co-infection. Co-infection with rotavirus and NoV was most common, and occurred in 20/155 cases (12·9%) including co-infection with adenovirus [Reference Koh22]. Information on the co-infection of NoV with V. cholerae O1 is limited from other parts of the world. In our study a significant number of NoV were co-infected with V. cholerae O1. Our study showed that in 22/78 cases, NoV alone accounted for the diarrhoea, with diarrhoea episodes ranging from 4 h to ⩾48 h with varying (n=2–12) number of stools per day. The incidence of NoV infection in children aged <5 years and adults was quite similar. All the patients had diarrhoea associated with some dehydration, two patients had fever and 16 (72·7%) patients showed no abdominal cramps or pain. Eighteen patients (81·8%) had >5 vomiting episodes in a 24-h period and two patients had no vomiting. Similar results were found in a study from Vilnius University Children's Hospital in 2005 [Reference Narkeviciute and Tamusauskaite23]. It was observed that in young children, NoV infection manifested as vomiting (94%), diarrhoea (81%), and fever (66%), and presented as gastroenteritis with fever (47%) or without fever (30%) [Reference Narkeviciute and Tamusauskaite23]. In our study, we found that six (7·3%) of 78 positive specimens belonged to NVGI strain and 72 specimens belonged to NVGII strains. Similar studies in the population of Cairo, Egypt, found predominance of GII.4 genotypes. The phylogenetic analysis of the capsid gene suggested that GII.4 strains from Cairo were similar to those circulating elsewhere [Reference Kamel24]. In our study we found six genotypes, i.e. GII.4, GII.13, GII.6, GII.1, GII.16 and GII.3; with predominance of GII.4 (n=8) and GII.13 (n=7) genotypes. In a study from Chiang Mai, Thailand, Khamrin et al. [Reference Khamrin25] observed that GII.4 was the most predominant genotype of NVs, followed by GII.15, GII.6, and GII.12. In a study from Nicaragua, nucleotide sequence analysis of NoV-positive samples of the N-terminal and shell region in the capsid gene revealed that at least six genotypes (GI.4, GII.2, GII.4, GII.7, GII.17, and a potentially novel cluster termed ‘GII.18-Nica’) circulated during the period of that study, with GII.4 virus being predominant [Reference Bucardo21]. An earlier study from Kolkata, found that 12 NoV cases (54·5%) were GII.4 and six cases showed 99% identity with the new variant Japanese strain Hu/NoV/GII.4/OC07138/JP, that study also detected three novel NVGII inter-genotype recombinant strains in children [Reference Nayak26]. In a study from Pune, India, it was shown that the phylogenetic analysis of partial RNA polymerase and VP1 (capsid) genes identified five NVGII strains (GII.4, GII.6, GII.7, GII.8, and GII.14 genetic clusters) with the possible occurrence of a ‘2007 new-variant’ of GII.4 [Reference Chhabra16]. A study from Delhi, India, found 36 (61%) positive for NoV (34 NVGII, 2 NVGI) and identified three genotypes (GII.4, GII.3, GII.b) in children with acute sporadic gastroenteritis [Reference Rachakonda27]. Our study reports first time on the appearance of GII.13, GII.6, GII.1, GII.16 and GII.3 NoVs in children and adults in Kolkata. From the present study, it is clear that NoV is an important viral aetiological agent having a significant role in gastroenteritis, in children and adults other than rotavirus in Kolkata. Therefore, routine surveillance for NoV infections in different settings and molecular epidemiological studies will provide interesting information that will enable us to understand the nature and spread of NoV infection.
ACKNOWLEDGEMENTS
We sincerely acknowledge the technical assistance of M. Hossain, B. Bera, K. Sen, A. H. Mallick, C. Bose and M. Guha and staff members of NICED Unit at Infectious Diseases & Beliaghata General (ID&BG) Hospital. We also sincerely acknowledge the invaluable support of our research trainees Parna Banerjee, Amrita De and Tanuja Khatun. This study was supported by a grant from the Okayama University Program of Founding Research Centre for Emerging and Re-emerging Infectious Disease, Ministry of Education, Culture, Sports, Science and Technology of Japan. The authors are grateful to Professor Y. Takeda and Dr G. B. Nair for their continuous support. M. K. Nayak, D. Chatterjee, S. M. Nataraju and M. Pativada were financially supported by fellowships from the University Grants Commission (UGC) India, Okayama University, and Indian Council of Medical Research (ICMR), respectively.
DECLARATION OF INTEREST
None.







