Improvements in the detection of enteric campylobacters and increased surveillance over the past four decades have shown that Campylobacter spp. are the most common cause of bacterial gastroenteritis in the developed world [Reference McClurg1]. Historically, the most prevalent enteropathogenic species have been the thermophilic C. jejuni (80–85%) and C. coli (10–15%) [Reference Moore2]. Although routine culture in the clinical laboratory favours the detection of C. jejuni and C. coli, several studies have demonstrated that campylobacters are capable of entering into a viable but non-culturable (VBNC) state upon exposure to adverse conditions such as sub-optimal nutrients, oxygen, osmotic pressure, temperature, and light [Reference Nogva3]. These bacteria fail to grow on routine bacteriological media. From a clinical perspective, when culture is the sole detection method employed, the consequence of the VBNC state leads to false negatives in the diagnosis of Campylobacter-related gastroenteritis. Another significant shortcoming of culture-based methods is that they generally fail to detect less common Campylobacter spp. including C. lari, C. upsaliensis, C. hyointestinalis and C. fetus, or Campylobacter-like organisms such as the recently re-classified C. ureolyticus [Reference Bullman4]. As a result culture-based detection is significantly biased towards the ‘common species’ such as C. jejuni and C. coli [Reference Moore2].
A recent study by O'Leary et al. [Reference O'Leary, Corcoran and Lucey5], in Southern Ireland, highlighted the potential limitations of routine culture methods by comparing conventional stool culture for the detection of enteric pathogens with an automated multiplex PCR-based system. The current study extends these findings and investigates the incidence of non-culturable Campylobacter spp. to determine the true prevalence of emerging campylobacteria which fail to grow under routine conditions in patients presenting with acute gastroenteritis.
Faecal samples from patients presenting with symptoms of acute gastroenteritis at Cork University Hospital, Ireland, between January 2009 and May 2010 were screened for Campylobacter spp. using the EntericBio® system (Serosep Ltd, Ireland); 436 samples were positive in this test. Samples were stored at 4°C and cultured with a sterile swab on modified charcoal cefoperazone desoxycholate agar (mCCDA; Oxoid, UK). The plates were incubated at 42°C in a microaerobic environment (CampyGen; Oxoid) and the presence of Campylobacter spp. was confirmed by characteristic microscopic and colonial morphology after 48 h. C. jejuni NCTC 11 322 type strain was incubated with each batch of plates as a positive control.
DNA was extracted from faecal samples, after overnight enrichment in the EntericBio broth, using the EntericBio system (Serosep) in accordance with the manufacturer's instructions and stored at −20°C until required. A total of 204 DNA samples were investigated using uniplex species-specific PCR assays for C. jejuni, C. coli, C. lari, C. fetus, C. hyointestinalis, C. upsaliensis and C. ureolyticus (Table 1). All PCR amplifications were performed in a mixture (25 μl) containing 3 μl template DNA, 12·5 μl 2× BioMix Red (MyBio Ltd, Ireland), 8·5 μl molecular grade water and 1 μl species-specific primer pair (25 pmol/μl). The cycling conditions were 35 cycles of denaturation at 95°C for 30 s, annealing for 1 min and extension at 75°C for 30 s. PCR amplicons were separated by electrophoresis in 1·5% agarose gels, stained with ethidium bromide and photographed under UV light. HyperLadder II (Bioline, MyBio, Northern Ireland) was used as the molecular weight marker. Control strains for PCR were obtained from DSMZ (Germany): C. jejuni subsp. jejuni DSM 4688, C. coli DSM 4689, C. lari subsp. lari DSM 11375, C. fetus subsp. fetus DSM 5361, C. upsaliensis DSM 5365, C. hyointestinalis subsp. hyointestinalis DSM 19053 and C. ureolyticus DSM 20703.
Table 1. PCR primers and annealing temperatures
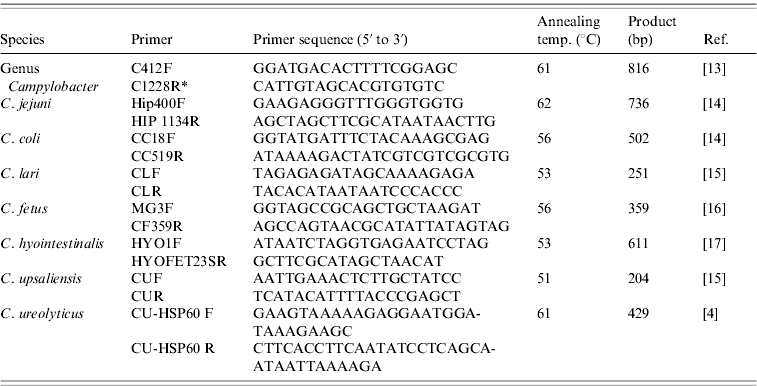
* Inglis & Kalischuk [Reference Inglis and Kalischuk17] reported a typographical error in the paper by Linton et al. [Reference Linton, Owen and Stanley13]: Primer C1288R should have read primer C1228R.
A Campylobacter genus-specific PCR targeting the 16S rRNA genes was performed on DNA samples negative by species-specific PCR. The PCR amplicons of 816 bp were DNA sequenced (Eurofins MWG Operon, Germany) and sequences were analysed by BLAST using the NCBI database and aligned by the ClustalW method in Megalign from the Lasergene suite of programs (DNAstar, GATC Biotech AG, Germany). Species identity was confirmed by total sequence similarities (GenBank) of ⩾99%.
Of 436 Campylobacter genus-positive patient faecal samples identified using EntericBio, 204 (46·8%) failed to yield campylobacters on culture. Furthermore, 20 samples positive by EntericBio tested negative in the species-specific PCR assays and of these 13 were negative in the Campylobacter genus-specific PCR and thus were considered to be false positives; the remaining seven samples were positive by the latter PCR but were not confirmed by 16S rRNA sequence analysis.
A total of 184 non-culturable samples were positive by both the EntericBio system and uniplex Campylobacter species-specific PCR, and including the seven samples above, give 191 samples. In total, 214 Campylobacter organisms were detected and included 21 patient samples with mixed infections involving 44 organisms. Figure 1 a shows that C. jejuni was identified in 97 (50·8%) samples and C. coli was relatively rare (5·7%); C. ureolyticus accounted for 80 (41·8%) of the samples. The most common species contributing to mixed infections were C. jejuni (17/21) and C. ureolyticus (15/21); two samples each yielded positive amplicons for three different species.
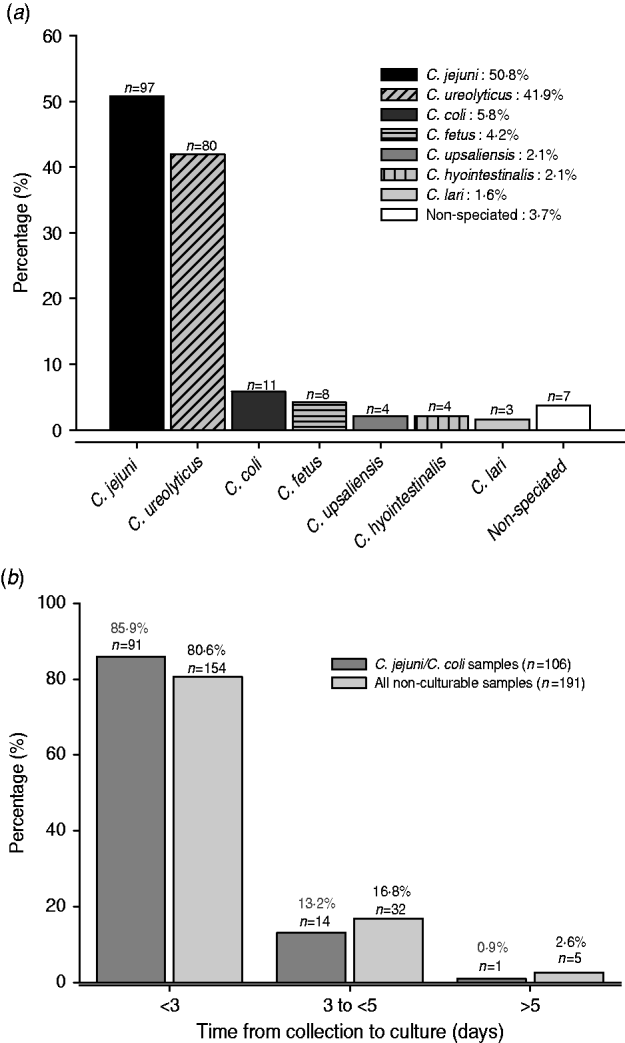
Fig. 1. (a) Species contributing to all non-culturable Campylobacter samples detected by the EntericBio system. Percentage of species detected relative to the total number of Campylobacter-positive patient samples (n=191). (b) Turnaround time from sample collection to culture for all non-culturable and C. jejuni/C. coli positive samples. Samples were stored at 4°C upon receipt at the clinical laboratory until time of culturing.
It was the intention of the study to culture faecal samples within 3 days of receipt of the specimen to maximize the recovery of Campylobacter spp., but a minority of samples were not cultured up to and including day 5. Figure 1 b shows that 85·9% of the 106 non-culturable C. jejuni/C. coli-containing samples were cultured within 3 days of receipt, a time frame currently accepted by routine practice. Moreover, 82 (42·8%) of the non-culturable samples yielded amplicons specific for species other than C. jejuni/C. coli/C. lari organisms which would not be normally detected by routine culture regardless of the time period from sample collection to culture. Indeed, the combination of the genus- and species-specific PCR assays confirmed that the EntericBio test system increased Campylobacter detection by 66·4% compared to culture within 3 days of sample collection.
For several decades C. jejuni and C. coli have been the prevalent species isolated from Campylobacter-related gastroenteritis in humans [Reference Moore2]. Developments in molecular diagnostics have, however, revealed that routine culturing methods favour the detection of these particular species over others and bias not only the diagnosis of Campylobacter-positive samples but also distort the contribution of other species to Campylobacter infections. The current study set out to investigate the observed discrepancy between culture- and molecular-based detection methods.
Compared to the preliminary findings reported by O'Leary et al. [Reference O'Leary, Corcoran and Lucey5] we detected a further 18·2% increase in the number of non-culturable Campylobacter genus PCR-positive samples. A likely explanation for the observed discrepancy is sample processing time. Unlike the earlier study [Reference O'Leary, Corcoran and Lucey5] in which samples were cultured on the day of receipt, here samples were stored at 4°C and cultured retrospectively once identified as Campylobacter-positive using the EntericBio method. In support of this, Ladron de Guevara et al. [Reference Ladron de Guevara6] previously reported that 16·2% of Campylobacter-positive samples failed to grow when comparing immediate culture with a 24-h delay stored at 4°C; a figure which aligns closely with our finding here. We found that 85·9% of the C. jejuni-/C. coli-positive samples which failed to grow were cultured within 72 h of collection (Fig. 1 b). This may reflect the fastidious nature of Campylobacter spp. and their adaptation to withstand the environment outside of the host's gut, i.e. entering into a VBNC state [Reference Nogva3]. Moreover, these findings raise significant concerns regarding delays in culturing protocols given that it is currently accepted practice for diagnostic hospital laboratories to refrigerate faecal samples received on a Friday for culture analysis on Monday; delaying culture by a further 48–72 h after collection. Additionally, the majority of faecal samples received are sent via general practitioners which further increases transit time from collection to receipt (without refrigeration) and ultimate culturing. It is, however, important to note that 42·8% of the non-culturable samples contained only Campylobacter spp. that are not readily detected by routine culture (i.e. non-C. jejuni/C. coli/C. lari) regardless of the time delay from sample collection to culture. While mCCDA was the selective medium of choice in the current study, as opposed to Preston agar favoured by O'Leary et al. [Reference O'Leary, Corcoran and Lucey5], it is unlikely that the media alone contributed to the observed discrepancy, at least for the ‘common’ thermophilic Campylobacter spp. given that isolation rates using mCCDA are similar, if not superior, to those using Preston agar [Reference Rodgers7]. Interestingly, in spite of the fact that routine Campylobacter culture methods favour the detection of C. jejuni, this species emerged as the most prevalent among the non-culturables and this might suggest transition to the VBNC state to enhance survival [Reference Nogva3]. In this state they are likely to have reduced metabolic activity and as a consequence are unable to utilize constituents of selective media. Campylobacter spp. are notorious for their fastidious in-vitro growth requirements despite their presence in nature in a variety of inhospitable environments [Reference Moore2]. Conversely, it is important to note that PCR assays detect a DNA target rather than intact live bacterial cells, and hence a proportion of the non-culturable C. jejuni may be dead or a previous Campylobacter infection. Additionally, as the EntericBio method incorporates an overnight enrichment step prior to DNA extraction it is likely that a proportion of these non-culturable samples contained levels of viable Campylobacter that were below the detection limit for mCCDA but following enrichment contained sufficient cell numbers for a positive PCR. A similar situation may also exist for the thermophilic C. coli strains identified in 5·8% of the non-culturable samples.
Approximately half of the species detected by PCR were non-C. jejuni/C. coli spp. with C. ureolyticus being the second most common. Despite its high incidence, current routine culture methods are inadequate for the isolation of C. ureolyticus as it requires an enriched hydrogen, microaerophilic environment with an optimum temperature of 37°C for growth [Reference Vandamme8]. The less common species (C. lari, C. fetus, C. upsaliensis, C. hyointestinalis), which collectively contributed to 10% of the non-culturable samples, are thought to be of particular clinical significance in immunocompromised patients [Reference Lastovica9]. Both C. fetus and C. hyointestinalis are non-thermophilic species and are incapable of growth at the 42°C incubation temperature used in the current culture-based methods [Reference McClurg1, Reference Mahar10]. Moreover, several reports have observed that the high concentrations of antibiotics used in Campylobacter selective media to suppress commensal gastrointestinal flora may also inhibit the growth of these less common species [Reference Mahar10]. Additionally, routine culture methods fail to provide the enriched H2 atmosphere required by C. hyointestinalis. In support of our findings Mahar et al. [Reference Mahar10] highlighted that direct culture using mCCDA was unable to detect at least one third of viable Campylobacter spp. from clinical specimens.
Although no current culture-based method is capable of isolating all Campylobacter spp., the Cape Town protocol has been noted as the optimal technique to recover most Campylobacter spp. from faecal samples [Reference Lastovica9]. Despite representing a significant improvement over existing culture methods, there are a number of limitations including a slow turnaround time, labour requirements in addition to its inability to isolate non-motile species or cells which have entered the VBNC state [Reference Lastovica and le Roux11]. The paucity of non C. jejuni/C. coli species reported to date, is likely due in no small part to the above-mentioned limitations and associated bias of existing culture techniques.
Mixed Campylobacter infections accounted for 11% of samples and C. jejuni and/or C. ureolyticus were present in all. It would be valuable to investigate if primary Campylobacter infection with a particular species increases the patients' risk of acquiring infection with a second ‘less common’ Campylobacter spp. or if both infections were acquired simultaneously from a particular source. It was noted, for example, that 75% of samples positive for C. fetus contained either C. jejuni or C. ureolyticus. Previous studies have found that C. fetus is a very rare cause of gastroenteritis in humans [Reference Lastovica9], yet we identified this species in 4·2% of the non-culturable samples which suggests that C. fetus, like C. ureolyticus, may well be an emerging human enteropathogen.
In conclusion, we report that the current ‘gold standard’ culture methods in the clinical laboratory are less than optimal for the detection of both the atypical and most common Campylobacter spp. in patient faecal samples. Our findings highlight that in addition to the underreporting of this statutory notifiable disease in Ireland [Reference Jansen12], routine culture (when performed within 3 days from sample collection) fails to detect over a third of Campylobacter-positive samples. This underlines the significant contribution of non-culturable Campylobacter spp. to human diarrhoeal illness and emphasizes the utility of rapid, sensitive and specific molecular detection methods to reveal otherwise undetected infections.
ACKNOWLEDGEMENTS
We acknowledge the assistance of Mr John Murphy, Department of Biological Sciences, Cork Institute of Technology. S.B. is supported by a scholarship from the Irish Research Council for Science, Engineering and Technology (RS/2009/1670). R.D.S. is an ESCMID Fellow. Funding was provided by Serosep Ltd, Ireland.
DECLARATION OF INTEREST
None.



