INTRODUCTION
The number of Salmonella enterica serovar Enteritidis infections has been declining in the UK since 1997 [Reference Lane1, Reference O'Brien2]. This decrease has been due to the successful adoption of vaccination and various other control measures in poultry production [Reference Hugas and Beloeil3]. However, at a European level, chicken eggs are still the most important source of foodborne Salmonella outbreaks [4]. Chicken eggs imported into the UK from other European countries have previously been associated with multiple Salmonella outbreaks [Reference Inns5–Reference Harker7].
Since April 2015, whole genome sequencing (WGS) has been used by the national reference laboratory in England and Wales as the routine procedure performed on all cultures of Salmonella sp. referred by local laboratories. WGS is the primary test for identification, surveillance and outbreak investigation by the reference laboratory; all other typing methods previously employed have been significantly reduced or withdrawn [Reference Ashton8, Reference Ashton9].
At the beginning of May 2015, an initial cluster of 29 S. Enteritidis cases was detected, and the cases were within a 5-single nucleotide polymorphism (SNP) single linkage cluster. These cases had specimens taken in a 2-month period; this was greater than the 10 cases within this 5-SNP cluster which were recorded in the whole of 2014. Twenty-three of the 29 cases were typed as S. Enteritidis phage type 59 (PT59). An outbreak was declared and Public Health England (PHE) led a multi-agency outbreak control team (OCT). The objective of the OCT was to identify the source of infection and implement control measures to prevent further cases.
METHODS
Case definition
Cases were ascertained from statutory notifications of cases of Salmonella infection. In the UK, local diagnostic laboratories are required to refer all Salmonella isolates to the national reference laboratories for confirmation and characterization. Cases were defined as persons resident in Great Britain and British Overseas Territories, with an onset of illness between 1 January and 27 November 2015, with S. Enteritidis infection which was whole genome sequenced and was within a specific single linkage cluster at the 10-SNP level. Cases were defined as travel associated if they had a history of travel outside the UK in the 7 days before onset of illness.
WGS
Sequencing was carried out by the PHE Genome Sequencing and Development Unit using Nextera library preparation and Illumina HiSeq 2500 (Illumina Inc., USA) in fast-run mode according to the manufacturer's instructions. High-quality Illumina reads were mapped to the S. enterica Enteritidis reference genome (GenBank accession no. AM933172) using BWA-MEM [Reference Li and Durbin10]. SNPs were then identified using GATK2 in unified genotyper mode [Reference McKenna11]. Genome positions that had a high-quality SNP (>90% consensus, minimum depth 10x, GQ ⩾ 30) in at least one strain were extracted and RAxML v. 8.1.17 used to derive the maximum-likelihood phylogeny of the isolates [Reference Stamatakis12]. Correlation between root to tip length and sample date was calculated using Path-O-Gen v. 1.4 (http://tree.bio.ed.ac.uk/software/pathogen/). Hierarchical single linkage clustering was performed on the pairwise SNP difference between all isolates at various distance thresholds (Δ250, Δ100, Δ50, Δ25, Δ10, Δ5, Δ0). The result of the clustering is a SNP address that can be used to describe the population structure based on clonal groups. FASTQ reads from all sequences in this study can be found at the PHE Pathogens BioProject at the National Center for Biotechnology Information (accession no. PRJNA248792), sample accession numbers are given in Supplementary Table S1. In addition to WGS, all isolates were phage-typed by the Salmonella Reference Service according to the method of Ward et al. [Reference Ward, De Sa and Rowe13].
Food chain investigation
Cases were interviewed using local questionnaires to ascertain foods eaten and other exposures in the 5 days before onset of symptoms. These questionnaires differed depending on where in the UK the case was interviewed. Shops, restaurants and other food outlets identified by cases were investigated by environmental health officers and the Food Standards Agency. Due to the previous associations between S. Enteritidis outbreaks and chicken egg consumption [Reference Inns5–Reference Harker7], food chain investigations concentrated on chicken eggs and chicken. From each point of sale, distributors and suppliers were traced by members of the OCT. Links between suppliers were mapped to produce a food chain network using yED Graph Editor v. 3.14 [14].
Network comparison
Potential links between the food chain network and the phylogeny were explored by comparing distances on the food chain network to genetic distances between isolates. Distances on the food chain network were measured as the number of intermediate nodes on the shortest path between pairs of cases (e.g. the distance between two cases infected at the same premises will be 1). Genetic distances between cases were computed as the patristic distances between the corresponding tips of the phylogeny. A Monte-Carlo Mantel test was used to assess the degree of correlation between the supply network and genetic distance matrices, using cases that were both sequenced and included in the food chain network; 9999 random permutations of the data were used to generate the null distribution of the Mantel correlation. Because such a relationship may be driven by cases infected by the same source, we also repeated this analysis using pairwise comparisons of cases with different sources of infection only. These analyses were carried out in R [15], using igraph [Reference Csardi and Nepusz16] for graph distances, adephylo [Reference Jombart and Dray17] for phylogenetic distances computations and ade4 [Reference Dray and Dufour18] for the Mantel test.
Food and environmental sampling
When our food chain investigations identified premises associated with several cases, environmental and/or food samples were taken, where possible. Food and environmental samples including raw shell chicken eggs (both shell and contents were analysed in pools of up to 10 eggs) were tested for Salmonella using the PHE national method FNES16 [19]. All presumptive Salmonella isolates were confirmed biochemically or by PCR and referred to the PHE Salmonella Reference Service for further confirmation and typing.
Spanish investigations
Due to epidemiological information which suggested a substantial proportion of cases had a history of travel to Spain, the National Centre of Microbiology, Institute of Health Carlos III, Madrid, Spain was contacted. Phage-typing is the primary method used by the Salmonella Reference Unit in Spain to type S. Enteritidis isolates. In Spain, 144 S. Enteritidis PT59 isolates were recorded in 2015, compared to an average of 28 in the previous 3 years. To investigate the link to cases in the UK, a convenience sample of 19 S. Enteritidis PT59 isolates (18 from human cases, one from a food sample) from 2015 were sent to PHE for WGS. Eleven of these isolates were from sporadic cases and eight were from two outbreaks linked to eggs; four isolates from humans in one outbreak, three human and one food isolate in the other outbreak.
RESULTS
Descriptive epidemiology
A total of 136 cases were identified, of which 21 were travel associated. Twenty-six (19·1%) cases were hospitalized. The cases' ages ranged between 2 months and 98 years (median 31 years); 24 cases were aged <10 years. Seventy-six (56%) cases were female. The first onset of illness was on 16 February 2015; the last date of onset was on 29 September 2015, the distribution of onset dates is shown in Figure 1. There appear to be two peaks of incidence, with the first peak at week 15, followed by a decrease and a second peak around week 35.

Fig. 1. Distribution of cases over time, by resident country of case. Specimen date used when onset date unavailable, 2015 (n = 40).
The geographical distribution of cases is shown in Supplementary Figure S1. The majority (90%) of cases were resident in England, 12 in Scotland and one in Wales. Non-travel-associated cases showed a strong degree of clustering around the centre of the UK; cases were most frequently from the West Midlands (n = 40) and North West (n = 33) regions. Of the 21 travel-associated cases, travel to Spain was reported by 12, travel to Portugal reported by three and two had links with Gibraltar; one travelled there and one was resident there. In total, 17 (81%) of the 21 travel-associated cases had a history of travel to Spain or the Iberian Peninsula in the 7 days before onset of illness.
Microbiological results
Phage-typing showed that 92 (68%) of the cases corresponded to PT59. Twenty-three cases were untypable, 10 were PT14b, five were RDNC (reacts but does not conform), three were PT56, two were PT62 and one was PT8.
Spanish isolates
All 18 isolates from Spanish cases were within the outbreak specific single linkage cluster at the 10-SNP level. For the convenience sample of Spanish cases; ages ranged between 3 and 104 years (median 44), 56% were female and specimen dates ranged between 8 April and 18 October 2015. The isolate from a food sample also fell within the outbreak-specific linkage cluster at the 10-SNP level. This food sample was from an omelette, sampled on 29 April 2015 as it was linked to three cases. It contained chicken eggs which were traced to a supermarket. Microbiological analysis of chicken eggs from the same lot was not positive for Salmonella spp. The eggs were traced to a farm in Spain which was inspected; this inspection did not indicate any infraction and microbiological investigation was negative.
Phylogenetic results
The outbreak forms a monophyletic cluster within the broader phylogeny of S. Enteritidis (Fig. 2). The nearest strain to the outbreak cluster (apart from previous cases, detailed below) was 11 SNPs away. The median pairwise distance between isolates meeting the outbreak definition was 10 SNPs (s.d. = 3·9 SNPs), with a maximum SNP distance of 20 SNPs (Supplementary Fig. S2); the phylogenetic tree (Fig. 3) also reflects this diversity. Within the five clusters where ⩾3 cases reported exposure at the same food establishment, the median pairwise SNP distance was 0 SNPs, with a maximum of 2 SNPs. Isolates associated with the Iberian Peninsula by residence or travel clustered throughout the tree. There was weak correlation between root to tip distance of the phylogeny and date of isolation (correlation coefficient = 0·53, Supplementary Fig. S3).
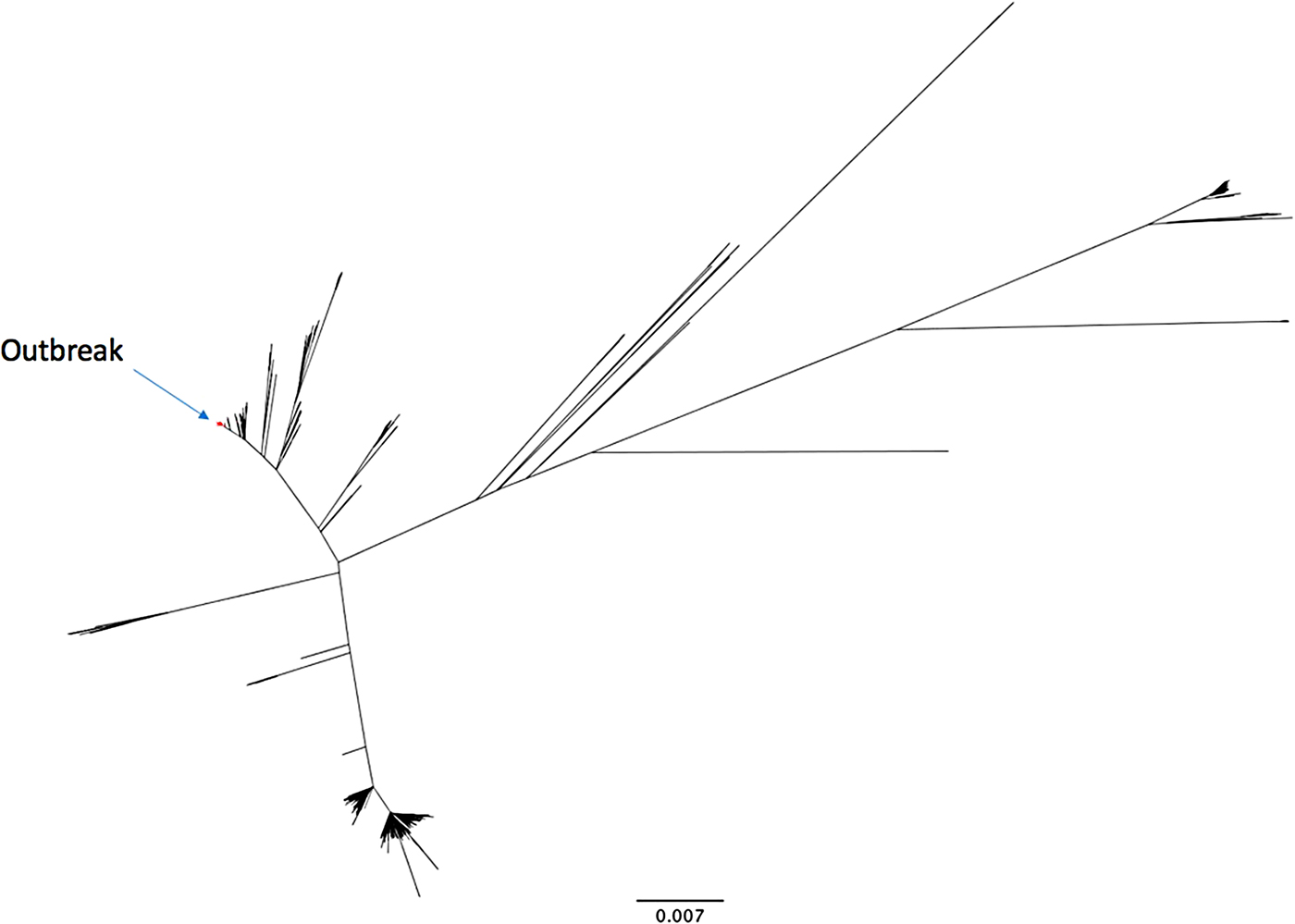
Fig. 2. An unrooted phylogenetic tree of 742 diverse Salmonella Enteritidis (eBurst group 4), built with 28 117 variant positions. The 136 outbreak isolates are highlighted in red.

Fig. 3. Maximum likelihood phylogenetic tree of the whole genome sequencing cluster containing the outbreak strains. Food isolate node label is highlighted in red.
Food chain investigation
Food exposure information was available for 87 (63%) cases. Of these, 63 (72%) had a history of eating out in the 5 days before onset. These cases visited 41 premises; chicken egg supply information could be obtained for 31 of these premises; 23 premises could be linked to a chicken egg supply network. The chicken egg supply network produced by this investigation is shown in Figure 4. Of the 87 cases with food exposure information, 43 (49%) cases could be plausibly linked to this network.

Fig. 4. Summary of egg food chain network, arrows show inferred direction of egg supply or exposure (total cases = 43).
Food and environmental sampling
A total of 17 catering premises, egg wholesalers and importers were sampled by environmental health officers from local authorities. A total of 1962 chicken eggs were sampled; none were positive for Salmonella sp. Twenty-six environmental samples were taken from catering premises; none were positive for Salmonella sp. Twelve chicken samples were taken; two raw chicken samples were positive for S. Livingstone and S. Enteritidis, respectively. The S. Enteritidis isolated from the chicken was >100 SNPs distant from the outbreak cluster, although by phage-typing the isolates were RDNC, which would not have allowed it to be ruled out as a causative agent without further typing. Six samples of other foods were taken; none were positive for Salmonella sp.
Network comparison
According to the Monte-Carlo Mantel test, genetic distances between isolates significantly increased with the distance (number of intermediate nodes) on the distribution network (r = 0·32, P = 0·0001) (Fig. 5). This relationship remained significant, albeit substantially weaker, when considering only pairs of cases from different origins (r = 0·07, t test: P = 0·03749), and was also robust to nonlinearities between distances (Spearman's ρ = 0·08, P = 0·02676).

Fig. 5. Jitter plot showing the relationship between the phylogenetic distance and the distance between cases on the food chain network, with cubic spline.
Previous cases
Sequences from isolates in this outbreak were uploaded to the GenomeTrakr database hosted at the National Center for Biotechnology Information (NCBI) [20]. An initial K-mer analysis was used to screen for similar isolates; these were then downloaded from the GenomeTrakr database and subjected to SNP analysis as described in the Methods section. SNP analysis showed that three cases from New York State were within the outbreak-specific single linkage cluster at the 10-SNP level. Follow-up information was only available for two of these cases. Both cases had onset of illness in 2014 and had a history of travel to Europe during their incubation period; one case visited Paris and one visited both Paris and London. In addition, there were 17 cases in residents of the UK with an onset of illness in 2014 and with a Salmonella isolate which was within the outbreak-specific single linkage cluster at the 10-SNP level (Fig. 3).
DISCUSSION
Here we describe an outbreak of S. Enteritidis in the UK that was linked to contemporaneous cases in Spain; these cases were within a specific single linkage cluster at the 10-SNP level. We found that cases were associated with chicken eggs; a food isolate within the outbreak cluster was taken in Spain from an omelette linked to several Salmonella cases. In addition, findings from food chain analysis for UK cases suggest that a number of these cases were linked by a chicken egg supply network; this network was significantly correlated with the phylogeny of the isolates. From this, we infer that UK and Spanish cases were exposed to a common source of Salmonella-contaminated chicken eggs.
This is the first published Salmonella outbreak to be prospectively detected using WGS. One of the main benefits of using WGS as the primary test for identification, surveillance and outbreak investigation is that it provides additional sensitivity over phage-typing (the previous routine typing technique for S. Enteritidis and a few other serovars) and far greater specificity in linking to isolates from other laboratories. Of the UK cases, 68% corresponded to PT59. Had WGS not been used, it is likely that the outbreak would still have been recognized, as phage-typing surveillance has previously been effective in detecting outbreaks [Reference Inns5]. The loss of 32% of cases would have likely slowed the recognition of the outbreak, and made it harder to pull together a food chain network, although whether the same source would have been identified is a matter of speculation.
An increasing number of countries are using WGS of Salmonella routinely for surveillance and outbreak detection [Reference den Bakker21, Reference Leekitcharoenphon22]. Due to information sharing and the use of databases such as GenomeTrakr, the ability to investigate multi-country Salmonella infections by comparing sequence information is greatly enhanced. Routine WGS also changes the way an outbreak is managed; it was previously accepted practice in infectious intestinal disease outbreaks to exclude cases with travel history to focus on possible in-country exposures. This practice does not preclude the inclusion of cases with foreign travel if the suspect source is from or in another country, but often the origin of the suspect source is not known. With the greater specificity of WGS information, travel histories and other geographical metadata can now provide information on which other countries may have cases from the same source [Reference Hoffmann23].
S. Enteritidis is considered a monomorphic pathogen that is hard to discriminate with previous typing methods (e.g. MLVA or PFGE) [Reference Zheng24, Reference Boxrud25]. Therefore, there may be an expectation that Enteritidis outbreaks analysed by WGS would form clusters with no detectable changes in their genomes. However, here we present an epidemiologically and genomically linked outbreak with considerable diversity, with a median SNP pairwise distance of 10 SNPs. We hypothesize that this is due to the outbreak being sampled from bacterial populations with a large effective population size, possibly due to contamination of multiple farms from the same source. The fact that isolates of this SNP cluster were also seen in 2014 is consistent with the source being chicken eggs, as laying hens can have a working life of 20 months (35 with moulting) [Reference Yousaf and Chaudhry26], and Salmonella contamination could conceivably persist in a flock over this entire period, or be transferred between consecutive flocks without detection [Reference Carrique-Mas27–Reference Davies, Wales and Sofos29]. We show that older isolates tended to be closer to the root of the tree, which is consistent with sampling from a persistent population over time. WGS also allowed the linking of three cases from 2014 from the United States as well, two of which were epidemiologically linked with Europe. This finding was facilitated by the GenomeTrakr project and New York State Department of Health colleagues, and is an example of the remarkable sensitivity and specificity of WGS. This type of international collaboration and comparison of WGS information provides exciting opportunities to enhance the safety of our global food supply [Reference Dunn30].
Despite the hypothesized association between these cases and chicken eggs, one of the limitations of this investigation is that we did not isolate Salmonella from any of the eggs sampled during this outbreak investigation. It has been previously documented that the time between chicken egg consumption and chicken egg sampling is typically greater than the shelf life of chicken eggs [Reference Inns5]. In this investigation we sampled 1962 chicken eggs and none were positive for Salmonella. The Spanish Reference Laboratory investigated 48 S. Enteritidis isolates from laying hens isolated during the salmonellosis control and prevention programme in 2015. PT59 was not detected in any of those isolates. It was, however, possible to isolate Salmonella in the outbreak cluster from an omelette in Spain, and there was a significant association between the chicken egg food chain and the genome phylogeny.
Food chain information was difficult to obtain in this outbreak; food exposure information was only available for 63% of cases; of 41 premises identified, egg supply information was obtained for only 31. This situation arose both from the challenges in contacting cases and poor recall, but also the very resource-intensive nature of traceback investigations to identify the food chain. In the European Union, food business operators (including importers) are legally required to ensure that on investigation, traceability can be assured at all stages (EC Regulation 178/2002). The experience of investigators in this outbreak was that this information was frequently unavailable and often very time-consuming to obtain.
Comparisons between the food chain network and phylogeny produced from WGS provide a promising new analytical application of this method for outbreak investigation [Reference Dallman, Inns and Jombart31]. Regarding the food chain information used in this analysis, it is important to remember that although local questionnaires asked similar questions, there could be variation between authorities in the questionnaires used and resources available for interview. If this difference in collecting information systematically varied across areas or countries, this could potentially affect the extent of the food chain network and any inferences drawn from comparisons using it.
In this outbreak, we have shown the way in which the routine use of WGS for Salmonella surveillance has provided additional opportunities for outbreak detection and investigation. The ability to collaborate on an international level in a timely and resource-efficient fashion would be enhanced by the use of an international database with good European and worldwide coverage. Ideally, this would include coverage of samples taken for farm and food monitoring, and for veterinary purposes. The greater use of WGS has also provided opportunities to develop new analytical methods to compare phylogenies with other sources of information for outbreak investigations. To make the most of these new methods, food chain investigations are of increasing importance, we recommend that national authorities work with food businesses to ensure they can meet legal requirements for traceability.
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268816001941.
ACKNOWLEDGEMENTS
This paper is written on behalf of the Outbreak Control Team; we acknowledge the contribution of all members, in particular Debbie Anderson for administrative support. We also acknowledge the work of environmental health officers in obtaining both information for the food chain investigation and food and environmental samples. We also thank Lesley Larkin and Rob Davies for advice on Salmonella in the poultry industry.
Thanks are also due to Bill Wolfgang and Madhu Anand from the New York State Department of Health for depositing their sequencing data and sharing their follow-up information. We also thank the GenomeTrakr initiative for organizing the data in such a way that it is possible to find closely related cases for further SNP analysis.
T.J. is funded by the Medical Research Council Centre for Outbreak Analysis and Modelling and the National Institute for Health Research – Health Protection Research Unit for Modelling Methodology. T.I., T.D. and R.V. are supported by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections in partnership with Public Health England (PHE). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, the Department of Health or PHE.
DECLARATION OF INTEREST
None.







