INTRODUCTION
Rotaviruses are a major cause of gastroenteritis in children aged <5 years worldwide [Reference Parashar1, Reference Parashar2] and of acute diarrhoea in the young of many other mammalian species (calves, piglets, lambs, rabbits, etc.). They cause more than 600 000 deaths each year, mostly of infants and young children in developing countries [Reference Parashar1], and are a significant cause of morbidity and hospitalizations in developed countries.
Rotaviruses are classified according to the serological cross-reactivity of the inner capsid protein VP6 into five groups (A–E) and two more groups (F, G) are likely to exist. Within group A rotaviruses, there are subgroups (I, II, I+II, non-I, non-II) characterized by their reactivities with two VP6-specific monoclonal antibodies [Reference Greenberg3]. More recently, RT–PCR has been applied to the subgroup classification of human rotaviruses, and only two major subgroups have been identified [Reference Iturriza Gomara4].
In order to characterize strains on the basis of their surface proteins VP4 and VP7, a binary classification scheme distinguishing types has been established for group A rotaviruses. The system differentiates G-types (VP7-specific, G for glycoprotein) and P-types (VP4-specific, P for protease-sensitive protein). To date, 23 different G-types and 31 P-types have been described [Reference Abe5–Reference Ursu9], indicating extensive genomic diversity within group A rotaviruses.
Rotaviruses possess a genome of 11 segments of dsRNA and VP4 and VP7 are coded for by different RNA segments (segments 4 and 7, 8 or 9 depending on strain, respectively). As rotaviruses were found to reassort readily in doubly infected cells in vitro and in vivo [Reference Schumann7, Reference Garbarg-Chenon, Bricout and Nicolas10–Reference Unicomb12], various combinations of VP4 and VP7 types have been observed in natural rotavirus isolates [Reference Esona13–Reference Matthijnssens16].
The European Rotavirus Network (EuroRotaNet), was established in January 2007 [Reference Iturriza-Gomara17], and has conducted rotavirus strain surveillance in Europe for three consecutive years. Participation in EuroRotaNet is voluntary, and the network activities are funded between the collaborating institutes and GlaxoSmithKline Biologicals (GSK) and Sanofi Pasteur-MSD (SPMSD). The participating institutes provide expertise, laboratory space, equipment and supervision, and funding for labour and consumable costs associated with strain characterization is provided in the form of an unrestricted collaborative grant in equal parts from GSK and SPMSD, and administered through the Centre for Infections, Health Protection Agency, London, UK.
The study was undertaken in order to gather comprehensive information of the rotavirus types co-circulating throughout Europe, including both urban and rural settings, and encompassing at least three consecutive rotavirus seasons. The aims of the study were (i) to develop methods and algorithms for effective rotavirus typing (G and P) and characterization (including VP6 and NSP4 genotypes) and to monitor the effectiveness of current genotyping methods and respond to changes associated with genetic drift and shift; (ii) to describe in detail the molecular epidemiology of rotavirus infections in Europe, during consecutive rotavirus seasons, through genotyping of rotavirus-positive samples collected throughout each country; and (iii) to monitor the emergence and spread of common and novel rotavirus strains within Europe and develop the infrastructure that may serve as a platform for future surveillance activities and nested studies. These studies will be used to monitor the effectiveness of a rotavirus vaccine in the general population, through monitoring the reduction in disease associated with common rotavirus types; to detect the possible vaccine-induced emergence of antibody escape mutants and the possible emergence in the general population of genotypes other than those included in the vaccine and reassortants between vaccine and naturally circulating wild-type strains.
MATERIALS AND METHODS
Samples
Hospitals, paediatricians and general medical practitioners that were willing to participate in the study over several years were identified in each of the participating countries in order to allow valid comparisons to be made between each rotavirus season and within each member country. Faecal samples submitted for the routine laboratory investigation of infantile gastroenteritis and positive for group A rotavirus antigen were collected for genotyping. A total of 19 140 rotavirus-positive samples were genotyped between September 2006 and August 2009 in 15 European countries (Table 1).
Table 1. Number of rotavirus strains in the EuroRotaNet database per country and rotavirus season (includes data for September to August in each of the three season from 2006 to 2009; entries for which date of sample collection was incomplete were excluded)
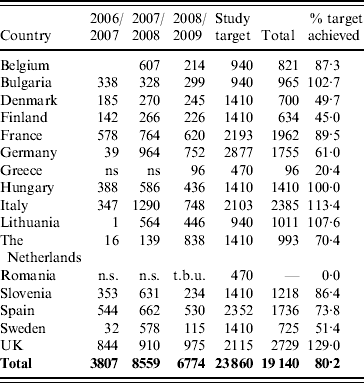
n.s., No sampling; t.b.u., to be uploaded.
Belgium, Bulgaria and Lithuania joined the network in 2008, Greece and Romania in 2009. Only data uploaded up to 31 December 2009 is included in the analysis.
Rotavirus genotyping
Rotavirus strains were characterized through genotyping using standardized methods to identify their G- and P-types [Manual of rotavirus detection and characterization methods, WHO (http://www.who.int/nuvi/rotavirus/WHO_IVB_08.17_eng.pdf); European rotavirus detection and characterization methods v4 (http://www.eurorota.net/docs.php)] and following the algorithm described in Figure 1. Rotavirus subgroups (VP6) and NSP4 genotypes were determined for uncommon or novel strains identified through G- and P-typing in order to identify possible zoonotic transmission [Reference Iturriza-Gomara18, Reference Iturriza-Gómara19] (European rotavirus detection and characterization methods v4).

Fig. 1. Algorithm describing the testing and reporting of rotavirus-positive samples.
Epidemiological data
Epidemiological data including age, sex, geographical location, including postcode in some countries, setting (hospital or community, urban or rural), symptoms (diarrhoea, vomiting, or diarrhoea and vomiting, or other) and date of onset and date of sample collection were entered into a web-accessible database and linked to the genotyping data (http://www.eurorota.net/).
RESULTS
Temporal distribution
Rotavirus infections peak in the winter months in temperate climates, and the analysis of the data in EuroRotaNet reflects this seasonality. The peak of rotavirus infections in Europe occurred in March in all three seasons between 2006 and 2009 (Fig. 2). The moving-average trend analysis indicated that rotavirus infections peaked in April in 2006/2007 and in March in 2007/2008 and 2008/2009 (Fig. 2).

Fig. 2. Temporal distribution of rotavirus infections in the EuroRotaNet database in three consecutive seasons, 2006/2007, 2007/2008 and 2008/2009. (a) Numbers of strains each month in three consecutive seasons. (b) Percent of the total rotavirus strains in a season each month showing the moving-average trend analysis.
Differences were observed across EuroRotaNet participating countries in the month in which rotavirus infections peaked. The earliest peaks of infection were detected in Spain in 2006/2007 and 2008/2009, in January and December, respectively, and in 2007/2008 infection peaked in February. Late season peaks were associated with countries in the East or North of Europe with infection peaking in Hungary in May 2007 and in Finland in May 2008 and 2009.
Genotype distribution
Total dataset
The total number of rotavirus-positive samples included were 19 140 for the 3 years and included 3807 in 2006/2007, 8559 in 2007/2008, and 6774 in 2008/2009. These numbers were used as denominators for calculating percentages. A total of 141 different combinations of G- and P-types were found between 2006 and 2009 and included those with combinations either singly or as multiple infections of G1, G2, G3, G4, G6, G8, G9, G10, G12 and P[3], P[4], P[6], P[8], P[9], P[10], P[11], P[14] genotypes (Table 2).
Table 2. Possible origins of rotavirus strains circulating within Europe. Common and reassortant human rotavirus strains, zoonotic strains and animal-human hybrid rotavirus strains are identified

Common rotavirus strains
G1P[8] rotavirus strains were predominant in all three seasons between 2006 and 2009. Strains circulating with a prevalence of >1% included G1P[8] (48·43%), G4P[8] (15·06%), G9P[8] (11·56%), G2P[4] (10·07%) and G3P[8] (4·27%) (Table 2).
Possible origins of rotavirus strains
Rotavirus strains found circulating within the European population between 2006 and 2009 can be stratified according to their possible origins. Human rotavirus strains are the most prevalent, making up 89·4% of circulating strains. Strains whose derivation may be associated with reassortment between animal and human strains make up 1·7% and reassortants of the common human strains make up 1·1%. Infection with rotavirus strains of animal origin may represent 0·3% of strains found (Table 2).
Mixed rotavirus infections
A total of 809 mixed rotavirus infections were detected between 2006 and 2009. These could be classified as single G-type with two or more P-types (148, 0·8%), multiple G-types with a single P-type (534, 2·8%) or multiple G- and P-types (127, 0·7%). These strains are representative of those genotypes seen in single infections and include mixtures of human and of animal rotavirus strains (Table 3 a–c).
Table 3 (a). Co-infections with multiple rotavirus strains (single G-type with two or more P-types)

Table 3 (b). Co-infections with multiple rotavirus strains (multiple G-types with a single P-type)
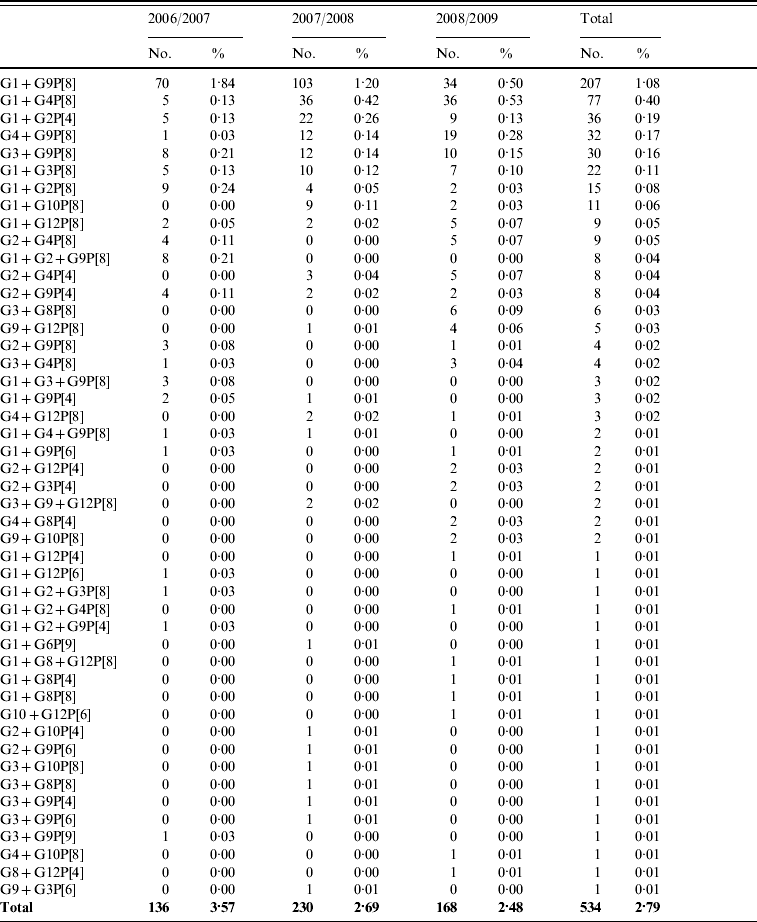
Table 3 (c). Co-infections with multiple rotavirus strains (multiple G- and P-types)

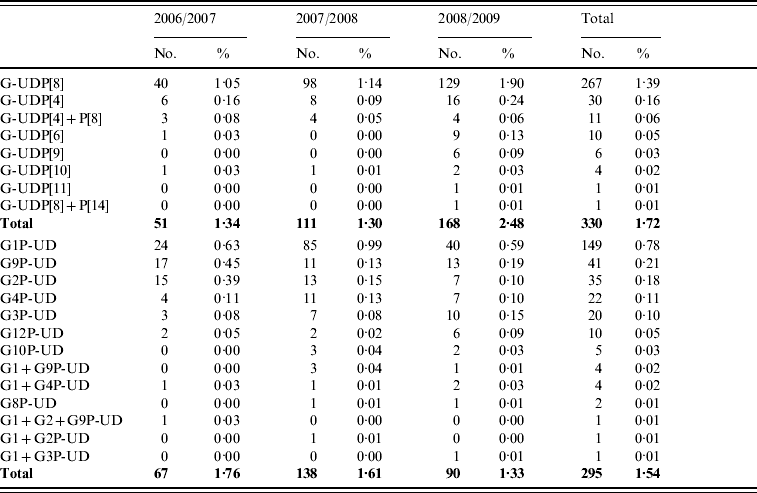
Partially typed rotavirus strains
A total of 625 strains were partially typed, in 330 (1·72%) and 295 (1·54%) strains, only the P-type or G-type, respectively, was obtained (Table 4).
Table 4. Partially typed rotavirus strains
Emerging rotavirus strains
Rotavirus strains emerging in Europe between 2006 and 2009 included G12 and G8 strains (Table 5). These genotypes were found circulating in each of the three seasons and both G12 (χ2=22·83, P<0·0001) and G8 (χ2=69·66, P<0·0001) showed significant increases in the third season.
Table 5. Distribution of possible emerging strains between 2006 and 2009 as single strain infections or contributing to multiple strain infections

Changing molecular epidemiology
G1P[8] strains were most prevalent in the 2007/2008 season with the exception of Lithuania where only 5·5% of strains were of this genotype. Over the three seasons, and in the majority of countries, the number of G1P[8] strains rose in the second season and fell in the third. The highest incidences of infection with G2P[4] strains in any season were in Belgium, Bulgaria and Greece where this genotype was found in >30% of the strains. In season 1, G2P[4] was the most prevalent strain in Bulgaria and in seasons 1 and 2 in Belgium. In the majority of countries, this genotype accounted for <20% of strains in any season. G3P[8] accounted for <20% of strains in all countries in any season. This strain was not found in the first season in Bulgaria, Italy or Hungary, in the second in Slovenia and in the third in Bulgaria and Greece. The prevalence of G4P[8] strains was 80% in Lithuania in 2007/2008 and >40% in Bulgaria and Germany in 2008/2009, and was found in all countries in the third season (2008/2009). G9P[8] strains were the most common strain found in Spain in the first season (2006/2007), and the second most common in Bulgaria, France, Italy and Finland (Fig. 3).
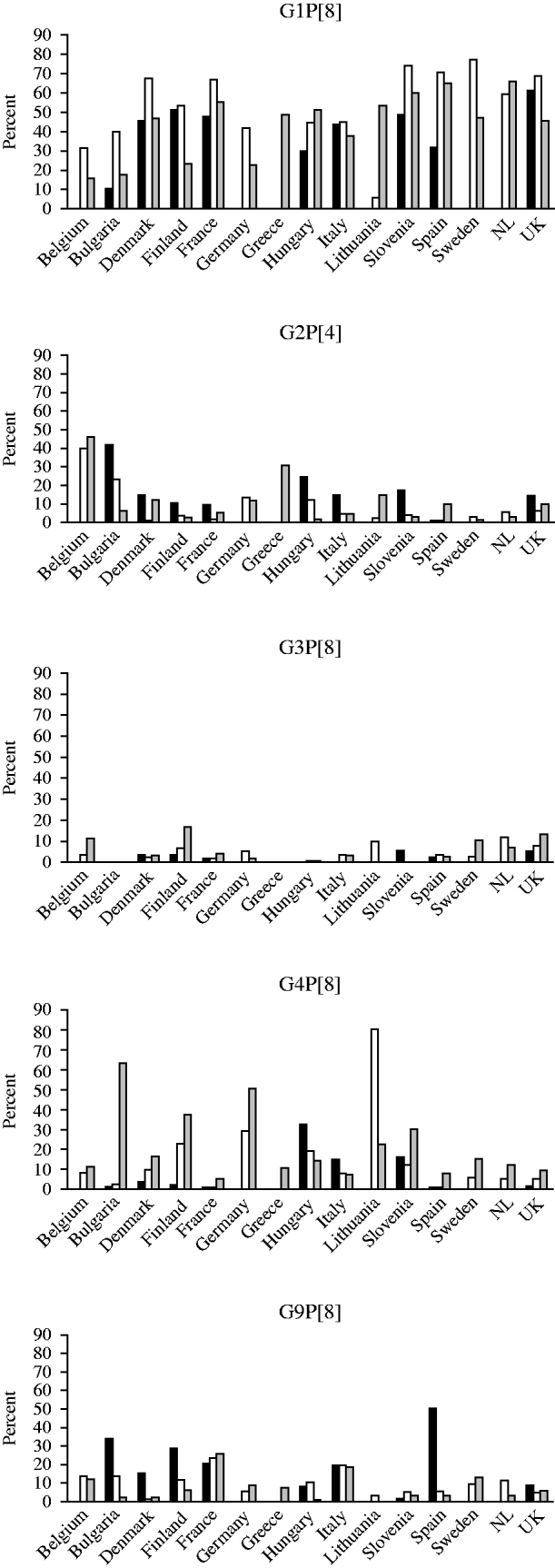
Fig. 3. Changing strain distribution in seasons and countries between 2006 and 2009. ▪, 2006/2007; □, 2007/2008; ![]() , 2008/2009.
, 2008/2009.
G12 rotavirus strains were found to be circulating with an incidence of >1% in nine countries including Finland (6·8%), Denmark (3·2%), Lithuania (2·4%), Hungary (2·3%), Greece (1·6%), The Netherlands (1·8%), Germany (1·7%), UK (1·5%) and Bulgaria (1·2%). With the exception of Sweden, G12 strains were found in all other countries. G12 strains were found in each of the three seasons in Denmark, Hungary, UK, Bulgaria, Italy and France.
G8 strains were found in two countries in the 2006/2007 season, in seven countries in the 2007/2008 season and in nine countries in the 2008/2009 season, with the highest incidence overall seen in UK (2·1%). The incidence was <1% in all other countries (Belgium, Bulgaria, Denmark, France, Germany, Greece, Italy, Lithuania, Spain, Sweden, The Netherlands).
Demographic data
Age distribution
Infection peaked in children aged 1–2 years, but was seen in all age groups (Fig. 4 a). Minor peaks, possibly associated with contact with infected children or grandchildren and waning immunity in the elderly population could also be seen (Fig. 4 b).
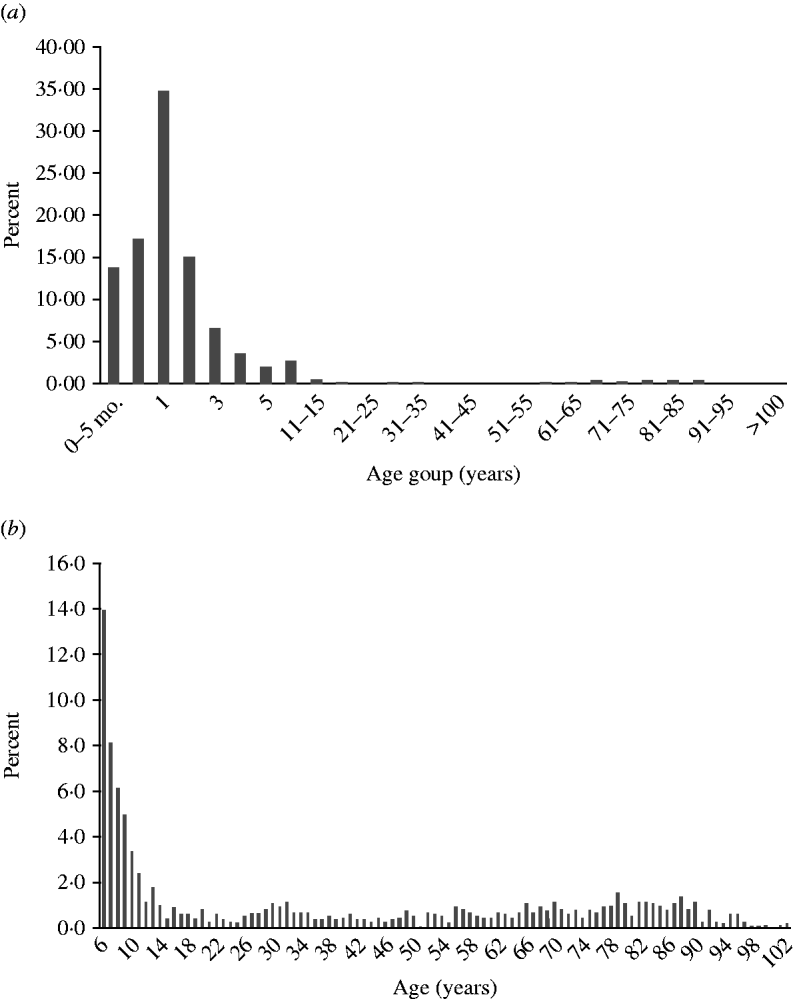
Fig. 4. Age distribution. (a) Rotavirus infections by age group in all countries (n=17 510). (b) Rotavirus infections from ages 6 to >100 years (n=1283).
Distribution according to hospital or community setting
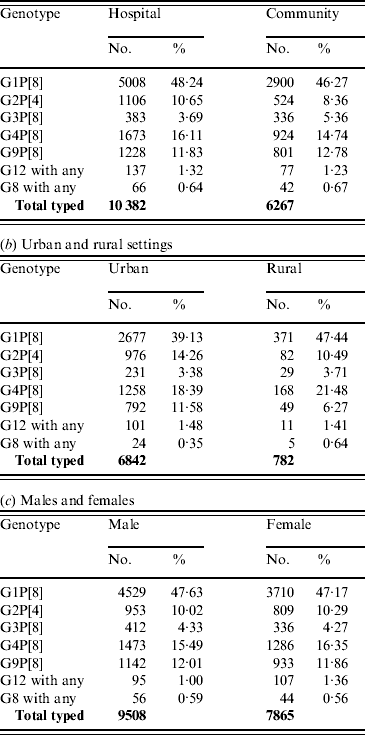
For those cases for whom the setting was provided, 62·4% of samples were collected from hospitalized patients and 37·6% from patients in the community. With the exception of G2P[4] strains there were no significant differences in the distribution of common or emerging genotypes between hospital patients and those in the community (Table 6 a). G2P[4] was significantly associated (χ2=22·98, P<0·0001) with hospitalized patients and may be associated with the periodic upsurge of G2P[4] strains, possibly associated with the selection of an antibody escape mutant. Infection can often require hospitalization as symptoms may be severe in the absence of cross-reactive antibodies associated with previous infections (see Age distribution section above).
Table 6. Distribution of common and emerging rotavirus genotypes. The total number of strains shown refers to the total number of rotavirus strains in each of the settings/gender groups, and includes mixed infections and human rotavirus reassortant strains (not shown in the table breakdown)
(a) Hospitalized and community cases
Distribution according to rural or urban setting
For those patients for whom the setting was provided, 89·7% samples were collected from urban populations and 10·2% from rural populations. The percentages of the European population living in urban and rural areas are estimated to be 73·1% and 26·9%, respectively (www.cap-lmu.de/fgz/statistics/urban_pop.php). G1P[8] strains (χ2=19·88, P<0·0001) were found significantly more often in rural populations, whereas G2P[4] (χ2=8·07, P<0·005) and G9P[8] (χ2=19·62, P<0·0001) were found more commonly in urban populations (Table 6 b). The increased incidence of G9P[8] and G2P[4] strains in the urban population may reflect the recent introduction of either an antibody escape mutant, such as seen previously with G2P[4], or an animal human reassortant, such as G9P[8]. The decreased incidence of G1P[8] in the urban population may be a result of the increased activity of G2P[4] and G9P[8] resulting in fewer susceptible individuals.
Distribution according to gender
For those subjects for whom gender was provided, 54·7% were male and 45·3% were female, which reflects the European birth cohort male:female ratio of 1·21:1. No significant differences in the distribution of common or emerging rotavirus strains were found between males and females (Table 6 c).
DISCUSSION
EuroRotaNet, was established in order to determine the diversity of co-circulating rotavirus strains in Europe over three or more rotavirus seasons from 2006/2007. Initially, 11 countries participated in the network and this had expanded to 16 countries by the 2008/2009 season. For analytical purposes, limited epidemiological data including age, sex, symptoms, date of onset, date of sample collection, country, region and settings, including hospital or community and rural or urban were collected.
The data on seasonality confirm the previously reported trend that rotavirus infections spread in Europe from South to North and West to East. This is similar to that observed in North America [Reference Turcios20].
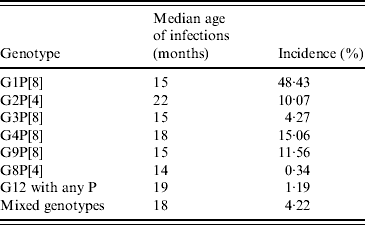
It might be expected that genotypes with a relatively low incidence are at a disadvantage in terms of transmission opportunities, and this is likely to be reflected by an increase in infections in older children. Data from Table 7, with the exception of G2P[4], do not support this, and the rationale for common strains such as G1P[8] and uncommon strains such as G8P[4] to significantly infect children between ages 14 and 15 months, is unexplained. The significant peak of infection in older children associated with G2P[4] may reflect poor cross-protection generated through previous infections with other common human rotavirus strains. Interestingly, this may reflect a replicative disadvantage of this strain in a mixture of predominantly fit viruses infecting an immunologically naive child. This taken with the development of immunological protection against the commonly circulating strains at a young age increases the probability that any new infection in an older child is likely to be with a less common, possibly less fit strain, such as G2P[4].
Table 7. Median age of infection compared with overall incidence by genotype
This report highlights the tremendous diversity of rotavirus strains co-circulating in the European population and points to the many origins of these strains including genetic reassortment and interspecies transmission. Mixed infections and zoonotic introductions are common, but many will lead to evolutionary dead-ends with only those virus strains demonstrating fitness for transmission within the human population emerging to become significant human pathogens.
The G- or P-types associated with partially typed strains were representative of those genotypes seen in fully characterized strains. Therefore, failure to type is most likely to be associated with technical issues and/or low viral load, although occasionally they may represent usual genotypes, particularly in those associated with an animal rotavirus G- or P- types [Reference Iturriza Gomara21–Reference Iturriza Gómara26]. The failure to identify G-types associated with P[6], P[9], P[10], P[11] and P[14] may be associated with the presence of unusual G-types for which G genotype-specific primers were not included in the study protocol. Similarly, the inability to determine the P-type of strains with G12 or G10 may be due to a lack of P genotype-specific primers. Interestingly, other strains associated with these G- or P-types were fully characterized during the study.
With the ability of the network to identify strains circulating with an incidence of 1%, possible emerging strains such as G8 and G12 have been identified during the first 3 years of the study and the analysis of recent data indicates their increased incidence.
G12P[6] and G12P[14] are likely to represent zoonotic spread into the human population, whereas, G12P[4] and G12P[8] are more likely to be the result of reassortment in human and animal strains. The plethora of mixtures of rotaviruses containing G12 strains may suggest environmental or food-/water-borne transmission events. Mixtures can also be seen to contain potential zoonotic strains other than G12 such as G8, G10, P[6], P[9] and P[11] and so may represent an environmental reservoir of rotavirus strains associated with a mixture of potentially contaminating human and animal faeces. Similarly, G8P[14] and G8P[6] may be zoonotic, whereas, G8P[4] and G8P[8] may result from reassortment of an animal strain carrying G8, with human strains. Twenty-four mixtures made up of 12 different combinations of G- and/or P-types were found, suggesting infection in some instances through contact with a mixture of human and animal strains.
The genes encoding VP7 of the G8 strains from the UK were sequenced. The temporal and geographical distribution and diversity of G8 strains seen before 2009 may suggest multiple zoonotic introductions [Reference Esona13, Reference Alfieri27–Reference Santos33] whereas the similarity of the strains described in the current paper is likely to be the result of sustained person-to-person transmission of a strain adapted to the human host (Fig. 5). Interestingly, when G12 strains from UK, Hungary and Bulgaria were sequenced, detailed phylogenetic analysis indicated multiple, unrelated, introductions and may represent a series of evolutionary dead-ends associated with an inability of these strains to sustain human-to-human transmission (Fig. 6). It is reasonable to suggest that because of the frequent introductions of G12 strains into the human population it may be only a matter of time before a reassortment event provides a G12 animal/human hybrid strain of comparable fitness to those current stains circulating commonly in the human population.

Fig. 5. Dendrogram (Neighbour Joining) of sequences of the gene encoding VP7 of G8 strains. Host, country and year of identification are indicated after each strain.
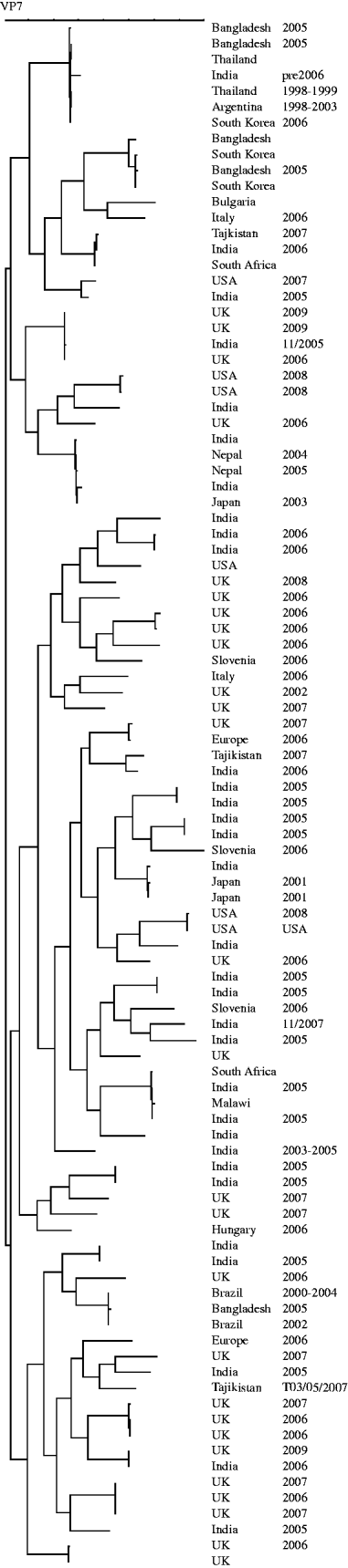
Fig. 6. Dendrogram (Maximum Parsimony) of sequences of the gene encoding VP7 of G12 strains. Strains included are global and represent introductions between 1991 and 2009.
The introduction of universal rotavirus vaccination in at least two of the participating countries, and partial vaccine coverage in some others may provide data on diversity driven by vaccine introduction and possible strain replacement. Discussions on changes to the current EuroRotaNet surveillance protocols in light of rotavirus vaccine introduction will be held during the course of the next two seasons and will involve participants, funders and the European Centre for Diseases Control (ECDC). In light of these developments, it should now be possible for EuroRotaNet, with others, to evolve into a network capable of performing detailed rotavirus surveillance activities. These activities would provide data on the effectiveness of rotavirus vaccination programmes and the impact of these programmes on the virus populations co-circulating in Europe, and the changing patterns of infections by age, season, setting, etc.
DECLARATION OF INTEREST
EuroRotaNet receives funding from GSK and SPMSD in equal parts through a non-restrictive collaborative grant.

















