INTRODUCTION
Escherichia coli are a normal constituent of the microflora of the gastrointestinal tract. However, certain E. coli types have been associated with disease in humans and animals. Based on their clinical features, virulence and adherence properties, E. coli strains associated with diarrhoeal disease have been subdivided into six different categories or pathotypes [Reference Nataro and Kaper1]. Verotoxigenic E. coli (VTEC) belong to one of the six pathotypes and are responsible for a large number of outbreaks of human diarrhoea ranging in severity from non-bloody diarrhoea to haemorrhagic colitis (HC) and haemolytic uraemic syndrome (HUS), a leading cause of acute renal failure in children [Reference Griffin and Tauxe2, Reference Karmali3].
VTEC O157:H7 is the most common serotype associated with HUS and HC, but it is far from being the only serotype associated with this disease [Reference Karmali3]. Serotypes such as O26:H11, O111:H8, O145:H28 and O103:H2 are among non-O157 serotypes frequently associated with HUS-related illness in humans [Reference Nataro and Kaper1], and are currently among the most prevalent VTEC serogroups in Europe [4]. In fact, over 200 E. coli [5] serotypes associated with verocytotoxin production have been isolated from humans. It has been estimated in the USA that 265 000 VTEC infections occur each year, of which only 36% are caused by O157 strains, with the remainder being caused by non-O157 VTEC. However, the numbers are likely to be higher due to the challenge in identifying non-O157 VTEC strains (http://www.cdc.gov/ecoli/general/index.html).
Although the main feature of any VTEC strain is the production of verocytotoxins, which are important in pathogenesis, a repertoire of virulence determinants has been shown to be associated with disease [Reference Nataro and Kaper1–Reference Karmali3]. A number of studies have used a polymerase chain reaction (PCR)-based binary typing method, whereby the presence of a gene is scored as 1 and absence as 0, to provide a genetic fingerprint of virulence genes present and a potential risk assessment of these strains to humans [Reference Coombes6–Reference Bugarel, Beutin and Fach8]. Others have used information on the ability of particular VTEC serotypes to produce epidemics and/or HUS disease in humans, in combination with the presence of virulence genes, to designate seropathotypes (SPTs) A–E; with strains from SPTA having most severe sequelae of infection for humans while strains from SPTE are usually confined to animals [Reference Karmali9]. In this study utilizing PCR we assessed the presence of 10 virulence genes for 365 E. coli strains belonging to O157 and non-O157 VTEC serogroups gathered from 11 European countries. The rationale for choosing these genes was based on their function (Table 1), which has been mainly associated with human illness or severity of clinical infections. Although the criteria for choosing these genes were not based on animal disease, virulence profile of VTECs isolated from diarrhoeic calves and other livestock have shown an association between the presence of virulence factors such as verocytotoxin, intimin and enterohaemolysin and disease [Reference Lee, Hur and Stein10–Reference Hutchinson12]. Therefore, the aim of this work was to use the PCR data and designated SPT to provide a potential molecular risk assessment of the individual strains and their ability to cause disease and/or epidemics.
Table 1. Sequence of the PCR primers and function of genes used in this study for virulotyping the E. coli strains

METHODS
Bacterial strains
A total of 365 strains were characterized. Most were chosen randomly from common serotypes representative of O157 and non-O157 serogroups associated with verotoxin production isolated from a variety of hosts from Denmark, France, Italy, Spain, and the UK; the O157 and non-O157 isolates from Sweden and Hungary, were representative of their prevalence in their respective countries (see Supplementary Appendix 1, available online). The number of isolates from each country were as follows: Austria (2), Croatia (2), Denmark (32), France (15), Germany (11), Hungary (50), Italy (92), Spain (19), Sweden (61) and the UK (England, Scotland, Wales, 81). There were 162 strains from serogroup O157; while the remaining 203 strains were from a number of non-O157 serogroups previously shown to be VTEC [Reference Scheutz13–Reference Orden19] (Supplementary Appendix 1) but did not include the O104 serotype as this study commenced before the German outbreak [Reference Mellmann20]. The strains were chosen prior to any knowledge of their virulence profile by PCR.
Growth, PCR amplification of virulence genes and analysis of data
Strains were cultured at 37°C, aerobically in Luria–Bertani (LB) broth for 16 h. Following culture, 1 ml cell culture was centrifuged at 16 000 g and the pellet resuspended in 300 μl TE buffer [10 mm Tris (pH 8), 0·1 mm EDTA]. For cell lysis the suspension was heated for 10 min at 95−100°C, cooled immediately and centrifuged at 16 000 g . The supernatant was collected and 5 μl used as a DNA template for PCR assays. The 10 genes included in this study are listed in Table 1; all PCRs were performed as simplex assays. Presence of the full efa1 gene was determined by two separate PCRs (efa1-5′ and efa1-3′).
PCR reactions were performed in a total volume of 25 μl containing 5 μl cell supernatant, 0·4 μ m of each primer (MWG Biotech, USA), 200 μ m of each dNTP, 2·5 μl × 10 PCR reaction buffer, 1·5 mm MgCl2, 0·75 U platinum Taq DNA polymerase (Invitrogen, USA). All 365 E. coli strains were screened with a panel of 11 PCR primer pairs representing 10 virulence genes selected to represent virulence characteristics associated with VTEC (Table 1). Results of the PCR performed for each strain are in listed in Supplementary Appendix 1 and are binary coded.
To find associations between strains, the binary-converted PCR data for all genes for the 365 strains were analysed in Bionumerics version 5.10 (Applied Maths, Belgium), whereby agglomerative hierarchical clustering was applied to the Euclidean distances between strains. The unweighted pair-group method with arithmetic mean (UPGMA) was used to construct the dendrogram. Strains were assigned to SPTs using previously published criteria [Reference Karmali9]; SPTA were O157:H7 or O157:H– serotypes which have caused epidemic HUS disease in humans; SPTB were non-O157 of multiple serotypes which have caused epidemic HUS disease in humans; SPTC were non-O157 of multiple serotypes which have caused sporadic HUS disease in humans; SPTD were non-O157 of multiple serotypes which have caused sporadic non-complicated diarrhoea in humans; SPTE were non-O157 multiple serotypes not associated with disease in humans.
Statistical analysis
Cramer's V statistic was used to determine strong association between each pair of genes/PCR for all 11 PCR products for serotypes O157 and O26. The two nominal variables were the genes/PCR products (Table 2) and each was a binary variable, i.e. had two levels, being present or absent. A Cramer's V statistic value of ⩾0·7 was chosen for a gene/PCR pair from within a country considered to be strongly associated. Such pairs were tabulated (Table 2) to compare virulence characteristics of strains for these serotypes.
Table 2. Associations between genes in E. coli serotypes O157 and O26 from different European countries calculated by Cramer's V association matrix. Only associations >0·7 are shown for simplicity*

* These may be positive or negative associations.
† Numbers in parentheses indicates total number of isolates from each country.
RESULTS
Differences in virulence genes present in O157 and non-O157 strains
In the present study, the presence of 10 virulence genes (Table 1) were chosen from those known to be involved in pathogenesis in VTEC, particularly O157:H7, encoded on integrated lamboid prophages, O-islands and plasmids [Reference Perna29], for screening the panel of E. coli strains. The resulting binary virulence typing or virulotyping data was analysed by hierarchical clustering (Fig. 1) and each of the main clusters was designated a SPT to give insight to the risk posed by these strains.
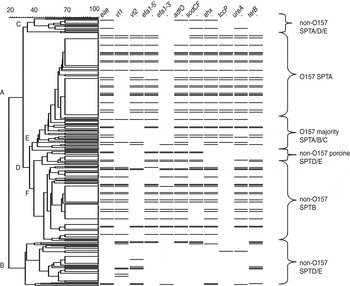
Fig. 1. The virulotype fingerprint for each strain and the resulting seropathotype (SPT) of the major groups identified from clustering of the binary-converted PCR data in Bionumerics (v. 5.0): A, B, C, D, E and F. Subclusters were designated as O157 or non-O157 depending on whether the number of O157 strains were the majority or minority in the cluster; and presence of the different serotypes was reflected in the SPT designation. Based on this there were two O157 subclusters (O157 SPTA and O157 majority SPTA/B/C). Details of the strain characteristics within each group are given in Supplementary Appendix 2.
Cluster E: the main O157 group
Most O157 strains were present in cluster E, which is a subcluster of cluster A of the dendrogram (Fig. 1). The virulence profile of the O157 strains were not entirely homogenous, but between 78% and 94% showed presence of: eae, vt2, efa1-5′, adfO, sodCF, ehx, tccP, ureA and terB genes; only 2% of strains were efa1-3′+, indicating the efa1 gene was mostly truncated; vt1 was present in 40% of strains, either singly or in addition to vt2. In fact, this was the dominant virulence profile of strains within the O157 SPTA group in subcluster E with >90% of strains harbouring the previously mentioned virulotype (Table 3). This group included strains of bovine and human origin of serotype O157:H7 or H– strains (where the H complex data was available). Nineteen of the 33 human strains were associated with disease. The O157 strains included all Swedish, most Spanish, Italian and UK strains but only five Hungarian strains from this study; one Croatian and one Austrian strain were also present (Supplementary Appendix 2). However, four of the 117 strains within this cluster were non-O157 (i.e. O176:H–, O5, O111, O145); but showed typical O157 features such as presence of the tccP gene in the former three strains, while the latter strain showed presence of the truncated efa1-5′ gene. These strains had been isolated from bovine, foodstuff and human strains.
Table 3. Prevalence of the virulence genes in each seropathotype (SPT) group identified within clusters of the PCR data

* dom is the abbreviation for group dominantly containing strains of O157 SPTA serogroup and a few strains from non-O157 sergroups belonging to SPTB and C.
Strains within the second O157 majority SPTA/B/C subcluster harboured a majority of O157 strains and a few non-O157 strains; together strains within this subcluster corresponded with serogroups belonging to SPTA, B and C [Reference Karmali9] and hence was named O157 majority SPTA/B/C (Fig. 1). It contained 34 O157 strains mostly of H7 or H– complex, of which 15 were from Hungary, nine from Italy, and ⩽5 strains from the UK, Spain, Denmark and Croatia. There were nine non-O157 strains from the following serogroups: O145, O128, O111, O115, O103, O49, and O26. Eight of the 12 human strains had caused disease; five were O157 strains and the remaining three were from serogroups O26, O128 and O145. All eight strains that had caused disease in humans harboured the vt2 gene, although this gene was also present in two human isolates not associated with disease. In fact there was no discernible pattern in the virulence genes tested in the eight isolates that had caused disease in humans from the remaining four that had not. This was a general trend that we noted in the virulence profile of all isolates that had caused disease in humans compared to those that had not but were of human origin; there was little difference between each group although the isolates causing disease showed slightly higher percentage of eae and vt2 genes (Supplementary Appendix 2). There was no dominant ‘virulence profile’ which defined this group although all strains harboured the eae gene, >80% harboured adfO, sodC, tccP, ureA, and terB genes; and the majority (>60%) were vt2 and ehx positive (Table 3). This is in contrast to the preceding group (O157 STPA; Table 3), where >90% of strains harboured vt2, ehx, and the truncated efa1 gene. In fact in several strains both vt genes were absent. Within the panel of vt-negative strains 11 were O157:H7 strains of bovine origin from Hungary, of which six were also negative for the efa1 gene (Supplementary Appendix 2).
Cluster C harboured strains from serotype/groups belonging to SPTA, D and E [Reference Karmali9], with O157 strains being the most numerous serogoup. However, only 7/21 strains were O157 strains. The incidence of disease in humans was low in this cluster with only 4/10 strains causing disease in humans and three of these belonged to the O157 H7/H– complex. All strains in this cluster harboured the sodCF gene and >70% harboured the eae, adfO, and ehx genes; none were terB positive, and prevalence of vt genes was low (Table 3). The non-O157 strains belonged to the following serogroups/types: O146, O145, O108:H9, O103, O56:H7, O28:H28, and O26. Strains from this cluster were of human, porcine or ovine origin with all eight porcine strains from Hungary, four of which had caused disease in pigs.
Cluster B contained only non-O157 strains with one exception; these were six O157 strains from Hungary of bovine origin and various H-types. These O157 strains were all positive for tccP and occasionally positive for the ureA genes but negative for all other genes including both vt genes and eae (Supplementary Appendix 2).
Cramer's V statistic was used to determine the within-country relationship of the virulence genes for O157 strains where enough data was available. This was also useful to determine genes that are likely to be associated in strains from different countries (Table 2). Cramer's V statistic showed that the 51 Italian strains were significantly different but harboured three perfect correlations indicating that Italian strains possessed ureA and terB in the presence of tccP. The 26 strains from Hungary were also very different but were highly likely to harbour adfO, sodCF, and ehxA when eae was present. A large proportion of O157 strains from Hungary did not harbour vt1, vt2, efa1-5′, terB or ureA genes. An association statistic could not be calculated for the 35 Swedish and 16 Spanish strains due to the highly similar virulence patterns they harboured. Therefore, based on the virulence genes used in this study, the O157 strains from Spain and Sweden showed both high within- and between-country (i.e. Sweden and Spain) similarity. Whereas the Italian and Hungarian strains showed very little between-country similarity but some within-country similarity.
Clusters B and F: the main non-O157 group
A defining feature of the non-O157 strains was the virtual absence of tccP (96% negative) and low prevalence of vt2 (32% positive); all other genes were present in ∼60% of isolates including the efa1-3′ component. All non-O157 strains within cluster F harboured eae, and tccP was virtually absent (Table 3). These strains were divided into two major subclusters based on the remaining genotype. Most strains (94/104) within non-O157 SPTB cluster F were from serogroups O111, O26 and O103 and of mostly H–, H11 and H2 complexes, respectively, collected from different regions in Europe. The remaining strains belonged to serogroups O145 (6); O157 (2); O121 (1) and O5 (1). More than 80% of strains representing the prevalent serogroups (O111, O26, O103), harboured genes for vt1, efa1-5′, efa1-3′, adfO, ehx, ureA and terB (Table 3). All strains except two were of human or bovine origin; 24/54 human strains and 5/45 bovine strains had been associated with disease.
In contrast <5% of strains within the non-O157 porcine SPTD/E group within cluster F harboured either vt or ehx genes but all were efa1-3′ and >80% adfO and sodCF positive. Fourteen of the 19 strains were of porcine origin from Hungary; six belonged to serotype O4/O123:H11, three to O49:H10/H–, three to O45:H11, one to O103:H11 and one to O56. Three strains were human (O157:H8, O111:H2, O128:H2) and two of ovine origin (O26, O56). Seventeen of the 19 strains in this group had caused disease in their host animal.
Forty-nine of the 61 strains within cluster B harboured the vt genes (one or both), the remaining virulence genes, including eae, were present in <26% of strains (Table 3). The SPT of this group was designated SPTD/E based on serotype/group [Reference Karmali9]. Twenty-one strains were of human origin and the remaining strains were of bovine (18), foodstuff (8), porcine (3), ovine (2), wild bird (1) and unknown (8) origin. Seven of the human and one of the porcine strains had caused disease.
Cramer's V statistic was used to detect frequencies of the virulence genes harboured by E. coli O26 strains from Italy and Sweden, as these datasets were large enough. The correlation matrix found six perfectly positive associations for the 17 Italian strains which indicated that in the presence of eae, genes such as ureA, terB and efa1-3′ were always present; these strains were also highly unlikely to harbour vt1 and vt2 together (Table 2). In contrast, all Swedish strains again showed nearly identical virulence characteristics (eae +, vt1 +, efa1 +, adfO +, ehx +, ureA +, terB +) but an association matrix could not be calculated. Therefore, as with the O157 strains, the Swedish O26 strains showed high within-country similarity, which was distinct from the Italian O26 strains for which only a subset of genes was associated with all strains. There was not sufficient data present for the other non-O157 serogroups for Cramer's V statistic to be applied.
DISCUSSION
A virulotyping approach based on the presence of 10 virulence genes was used to assess whether this may indicate the potential risk posed to humans by 365 strains of both O157 and non-O157 serogroups. The cluster analysis clearly divided O157 and non-O157 strains into separate clusters based on the virulotyping data, which matched well with SPT designations, e.g. the ‘high-risk’ O157 SPTA strains harboured most of the chosen virulence genes except efa1-3′, while most of these virulence genes were absent from the ‘low-risk’ non-O157 SPTD/E strains from cluster B.
However, our virulotyping data show that there is a lack of gradation if only the seropathotyping scheme is used. For instance the ‘low-risk’ non-O157 porcine SPTD/E strains from cluster F were not distinguished from the non-O157 SPTD/E strains in cluster B, which were also identified as low risk, although they harboured far fewer virulence genes with strains from a variety of human and animal sources, of whom only a few were ill. Interestingly, about 90% of strains within the ‘low-risk’ non-O157 porcine SPTD/E cluster had caused disease in pigs compared to 50% of porcine strains in cluster C, designated as non-O157/O157 SPTA/D/E, which included some ‘high-risk’ strains from humans and hence received the SPTA designation. Therefore, the SPT designation although relevant to humans may be less relevant to other animal species. Porcine strains from both groups were from Hungary and a feature distinguishing the non-O157 porcine SPTD/E cluster was the presence in most strains of terB, part of the tellurite-resistant operon [Reference Taylor28] and complete efa1, the enterohaemorrhagic E. coli adherence factor [Reference Perna29]. The efa1 gene is present on the pathogenicity island termed OI-122, in the EHEC O157 reference strain EDL 933, whose presence has been shown to correlate with severe disease in humans [Reference Morabito21]; this data indicates it may also affect pigs.
The SPTA group, the main O157 group, contained O157:H7/H– strains gathered from several different regions of Europe which showed very similar virulence profiles. Their genotype was identical to the EHEC O157 strain reference strain EDL 933 [Reference Perna29]. All Swedish and most Spanish O157 strains were within the O157 SPTA cluster; they were also found to be highly homogenous using Cramer's V statistic indicating that they share this subset of important virulence genes, which could not be used to distinguish them further. Although most strains included were randomly chosen, the Swedish and Hungarian O157 strains were from previous studies [Reference Eriksson30, Reference Toth31], with pulsed-field gel electrophoresis (PFGE) data available, and reflected their prevalence in the respective countries. The PFGE data available for the Swedish strains showed that they could be divided further into subgroups with different profiles that have been linked with clinical outcomes [Reference Eriksson30]. In contrast the virulotype data of the Hungarian O157 strains, which were shown to be particularly heterogeneous from the clustering and Cramer's V statistic, showed a good match with the heterogeneity of the PFGE profiles [Reference Toth31]. Clustering from the PCR virulotyping had placed these O157 strains into three different clusters. Strains from cluster E SPTA/B/C could be matched to PFGE cluster A; from cluster E SPTA to PFGE clusters B/C; and cluster B non-O157 SPTD/E to PFGE cluster D. Therefore, although the 10-gene virulotyping scheme was discriminatory for the latter group, its level of resolution will need to be improved for highly homogenous strains, e.g. the Swedish O157. This could be done by increasing the number of target genes, as shown by Brandt et al. [Reference Brandt7] who used a 41-gene virulotyping scheme. Even though their data also showed PFGE to be more discriminatory, the PFGE clustering generally corresponded with the PCR virulotyping data.
Recent whole genome sequence analysis of selected strains from non-O157 VTEC serogroups (O26, O111, O103), has shown that they share many virulence genes that are found on prophages and plasmids in O157, which are acquired through horizontal transfer of the mobile elements [Reference Ogura32]. Our PCR results indicate that presence of these virulence genes is widespread in strains from these serogroups, which are known to be high risk to humans. We were not able to distinguish between strains only on the basis of human disease as strains with the same virulence genes showed different clinical outcomes (Supplementary Appendix 2), possibly indicating that manifestation of human disease is complicated by other factors such as host immune response. However, there was higher prevalence of vt2 genes than vt1 in the O157 group compared to the non-O157 strains of SPTB, which was reflected in disease occurrence: ∼60% of human strains within O157 (cluster E) caused disease while ∼42% of human strains within the non-O157 SPTB cluster caused disease. Another characteristic seen in the non-O157 group was the low prevalence of the tccP gene, which was expected, although it was lower than previously reported (14% [Reference Garmendia26]) and could be due to only the 1014-bp PCR amplicon being considered. We also observed that there were some differences in the virulence genes harboured by non-O157 strains that were present in cluster E (O157 majority SPTA/B/C) compared to those in cluster F (non-O157 SPTB); notably strains in the former group mainly harboured the truncated efa1 gene and none harboured the vt1 toxin, being more similar to cluster E O157 strains (Supplementary Appendix 2). It should be noted that strains in cluster B also harboured one or both vt genes but showed much lower incidence of disease in strains of human origin, indicating that other virulence factors are required in combination with vt genes to cause disease in humans.
The routine use of a simple PCR scheme and SPT for surveillance, as presented here, although less sensitive than PFGE, nevertheless correctly distinguished high-risk E. coli pathogenic strains from low(er) risk ones and is much simpler to perform. Therefore, on performing PCR, isolates showing virulence profiles similar to those present in cluster E or cluster F non-O157 SPTB (Table 3), would be expected to be high risk to humans while those with similar profile to cluster B isolates would be expected to be low risk to humans. The resolution of the PCR scheme can be improved if larger numbers of genes are used to virulotype isolates, which need to be small enough to be routinely practicable in reference laboratories. An alternative would be to use the 10-gene PCR virulotyping scheme presented here to identify ‘high-risk’ strains rapidly, which can then be followed by more discriminatory methods such as the use of larger numbers of virulence gene PCRs or the virulence gene array [Reference Anjum33], or even performing PFGE for further subtyping of high-risk strains to enable epidemiological tracing and risk analysis following outbreaks. The use of PCR to distinguish between high- and low-risk strains is not new; a pentaplex PCR panel has been successfully established for routine laboratory use as the minimal predictor of avian pathogenic E. coli which may be of high risk to both poultry and humans [Reference Johnson34].
SUPPLEMENTARY MATERIAL
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268813001635.
ACKNOWLEDGEMENTS
The authors acknowledge the Med-Vet-Net network of excellence (WP26) for funding the studies. The authors are grateful to Mr Mark Saunders for his contribution in consolidating the PCR data and the E. coli Reference Laboratory at the Animal Health and Veterinary Laboratories Agency for contributing strains for this study.
DECLARATION OF INTEREST
None.






