Impact statement
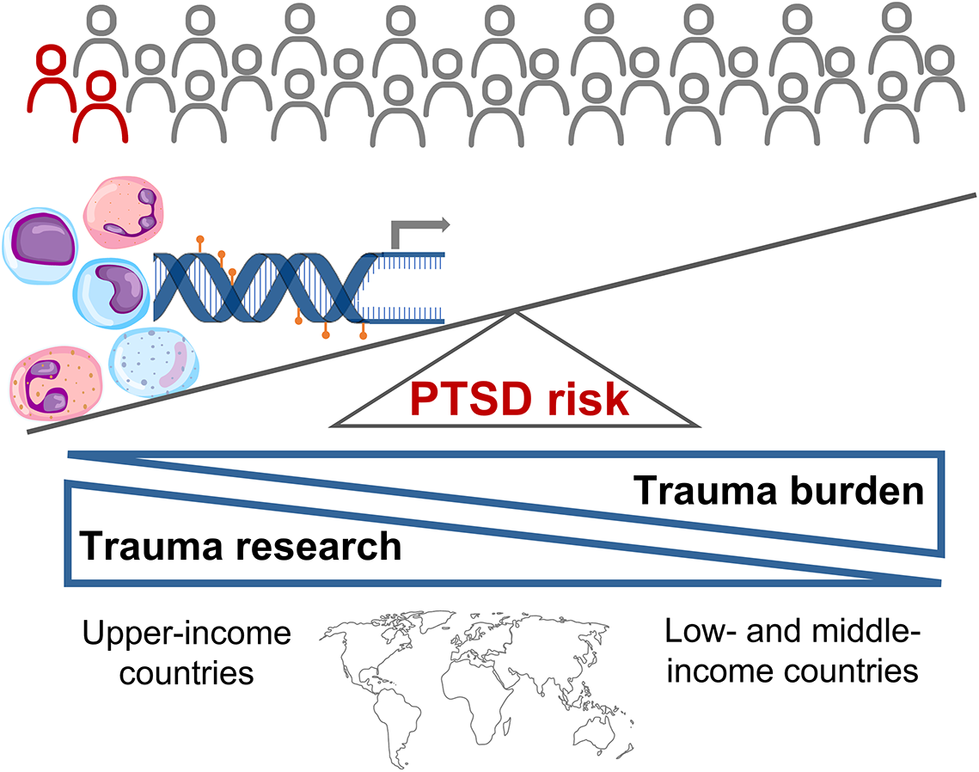
Though most people experience at least one traumatic event in their lifetime, only a subset go on to develop PTSD. Biological mechanisms play an important role in determining risk and resilience. Advances in molecular technologies and data analytic procedures now provide unprecedented insights into the molecular aetiology underlying PTSD. Ideally, identification of biological correlates of PTSD should be used to stratify trauma-exposed individuals according to risk, target preventative measures and interventions accordingly, identify biological targets for therapeutic modulation and track treatment response. This systematic review provides a synthesis of the evidence base globally, highlights key mechanisms and approaches used in molecular research, and compares and contrasts the nature and form of the literature base in upper- and low- and middle-income countries. Studies drawing on different biological markers, including genotypic, epigenetic, transcriptomic, endocrinological and serum level data, consistently point to the role of the stress response, inflammation, and learning and memory in PTSD symptomology. Contingent risk granted by these mechanisms depends on environmental and demographic factors. Our search results indicate a mismatch between the global trauma burden and the research conducted, with studies primarily conducted in upper-income countries. Detailed investigations of the molecular mechanisms underlying PTSD in diverse populations and contexts are required if the promise offered by biological insights is to be globally relevant, actionable and equitable.
Introduction
Trauma exposure is prevalent worldwide with approximately 70% of people reporting exposure to at least one traumatic event in their lifetime (Benjet et al., Reference Benjet, Bromet, Karam, Kessler, McLaughlin, Ruscio, Shahly, Stein, Petukhova, Hill, Alonso, Atwoli, Bunting, Bruffaerts, Caldas-de-Almeida, de Girolamo, Florescu, Gureje, Huang, Lepine, Kawakami, Kovess-Masfety, Medina-Mora, Navarro-Mateu, Piazza, Posada-Villa, Scott, Shalev, Slade, ten Have, Torres, Viana, Zarkov and Koenen2016). Based on analysis of 26 population surveys, around 5.6% of trauma-exposed individuals will go on to develop posttraumatic stress disorder (PTSD), which is characterised by symptoms of hyperarousal, reexperiencing, avoidance and negative alterations to cognition and mood (American Psychiatric Association, 2013; Koenen et al., Reference Koenen, Ratanatharathorn, Ng, McLaughlin, Bromet, Stein, Karam, Ruscio, Benjet, Scott, Atwoli, Petukhova, Lim, Aguilar-Gaxiola, Al-Hamzawi, Alonso, Bunting, Ciutan, de Girolamo, Degenhardt, Gureje, Haro, Huang, Kawakami, Lee, Navarro-Mateu, Pennell, Piazza, Sampson, Ten Have, Torres, Viana, Williams, Xavier and Kessler2017). Though PTSD contributes to poor mental health globally, low- or middle-income countries (LMICs) are disproportionately affected. Not only is the total population of LMICs substantially higher than that in upper-income countries (UICs), but these regions also suffer from a dual burden of potent stressors and limited mental health care resources (Purgato and Olff, Reference Purgato and Olff2015). For example, the age-standardised prevalence of PTSD in conflict settings is approximately 15.3% and this burden is primarily in LMICs (Charlson et al., Reference Charlson, van Ommeren, Flaxman, Cornett, Whiteford and Saxena2019). Hoppen et al. estimated that in 2019, more than 99% of adults who had experienced war in the preceding 30 years resided in LMICs, which by extrapolation accounts for approximately 3.1 million PTSD-associated disability-adjusted life years in these countries (Hoppen et al., Reference Hoppen, Priebe, Vetter and Morina2021). In addition, factors, such as higher socioeconomic status, living standards, community infrastructure and use of mental health services, which protect against the development of posttraumatic stress (PTS) following disasters and pandemics may be less prevalent in LMICs (Newnham et al., Reference Newnham, Mergelsberg, Chen, Kim, Gibbs, Dzidic, DaSilva, Chan, Shimomura, Narita, Huang and Leaning2022).
Neurobiology of traumatic stress outcomes
Biological mechanisms grant contingent risk or resilience and contribute to the considerable interindividual variability in posttraumatic stress (PTS) symptom presentation, severity, trajectory and treatment response (Ressler et al., Reference Ressler, Berretta, Bolshakov, Rosso, Meloni, Rauch and Carlezon2022). Translational neuroscience provides evidence for genomics, neural circuitry and neurotransmission aberrations and dysregulated immune system processes as causes or correlates of PTSD. PTSD is unique among disorders in requiring a precipitating traumatic exposure, which provides an opportunity for early preventative or ameliorative interventions. Identification of biological correlates of PTSD symptom trajectories has the potential to elucidate underlying biological mechanisms, stratify individuals according to relative risk, target and track the efficacy of treatment, and enable more accurate assessment of novel intervention strategies (Schultebraucks et al., Reference Schultebraucks, Sijbrandij, Galatzer-Levy, Mouthaan, Olff and van Zuiden2021).
Aims of the systematic review
The aim of this systematic review is to summarise recent insights into the molecular aetiology and pathophysiology of PTSD, identify key biological signatures involved, highlight methodological advances, and provide an overview of the global nature, state and representation of the research field. We focussed on longitudinal studies, as they can more accurately capture the complex and dynamic relationships between biological signatures and symptom trajectories and can assess the role of potentially modifiable factors as moderators or mediators of risk. To provide a more in-depth review, we additionally chose to focus on studies examining biological correlates as predictors of PTSD risk or symptom trajectory, rather than treatment response.
Methods
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines were employed in conducting the systematic review (Page et al., Reference Page, McKenzie, Bossuyt, Boutron, Hoffmann, Mulrow, Shamseer, Tetzlaff, Akl, Brennan, Chou, Glanville, Grimshaw, Hróbjartsson, Lalu, Li, Loder, Mayo-Wilson, McDonald, McGuinness, Stewart, Thomas, Tricco, Welch, Whiting and Moher2021).
Search strategy
Two researchers, JSW and MdP, independently searched the Pubmed, Scopus and Web of Science databases on 9 September 2022 for manuscripts published since 1 January 2021 using the following search terms: (("post-traumatic stress" (All Fields)) OR ("posttraumatic stress" (All Fields)) OR (PTSD (All Fields))) AND ((neurobiolog* (All Fields)) OR (genom* (All Fields)) OR (DNA (All Fields)) OR ("stress hormone" (All Fields))) AND (English(Language)) NOT (animal). These terms were designed to capture studies examining molecular correlates (including genomic, endocrine and inflammatory measures) of PTSD in human participants. The term ‘trauma’ was not included due to its high overlap with studies in the fields of surgery, physical rehabilitation and orthopaedics, which yield datapoints outside the scope of the review. The search was repeated using the same strategy on 22 March 2023 to include articles published up until 31 December 2022. This two-year time frame was chosen to focus on recent findings so that the review provides state-of-the-art insights into PTSD.
Eligibility criteria and selection process
The online open-source software Rayyan was used to collate the database search outputs, scan for duplicates and conduct a preliminary selection of articles (Ouzzani et al., Reference Ouzzani, Hammady, Fedorowicz and Elmagarmid2016). Suspected duplicate articles flagged by the software were manually checked and removed if necessary. Selection of publications was performed by applying inclusion and exclusion criteria to the study abstracts. Studies to be included had to (a) comprise original research manuscripts or meta-analyses of (b) cohort investigations that (c) assessed longitudinal relationships between (d) one or more molecular-level measures and (e) either PTS symptoms or PTSD status, and (f) be written in English. Exclusion criteria related to the manuscript type and research approach employed. Narrative reviews, case studies and protocol papers, as well as studies of exclusively healthy participants exposed to an experimental stressor or using animal models were excluded. The final study selection was based on a review of the full text manuscripts. A third researcher was approached when inclusion decisions were discrepant, and a final decision was reached by consensus. A total of 18 manuscripts were selected for the systematic review (Figure 1).

Figure 1. Study selection flow diagram. Molecular biology and PTSD.
Quality assessment
The quality of each study was independently assessed by researchers (JSW, MCG, MdP) using the Systematic Appraisal of Quality in Observational Research (Ross et al., Reference Ross, Grigoriadis, Mamisashvili, Koren, Steiner, Dennis, Cheung and Mousmanis2011). This tool evaluates the quality of evidence-based cohort and case–control observational studies in psychiatric research and assesses quality across five categories, namely sample, control/comparison group, measurement and output quality, follow-up, and distorting influences. The extent to which each study complies with the two to five statements listed under each category is used to classify quality as adequate, unclear or inadequate. A final quality level ranging from low to high was assigned based on these metrics.
Data extraction
Researchers (JSW, MdP, MCG) individually extracted data from articles using a template designed to capture information relevant to the review aims. The study design and setting category reports on the design, location of the study and/or site of participant recruitment, and the name of the parent study or cohort if applicable. The sample characteristics category records the number, sex, age and ancestry of participants broken down according to experimental group or study cohorts if necessary. The third category for data extraction was the overall stated aim of the study. Clinical measures record the PTSD assessment instruments, while the type of molecular measure and source tissue were included under molecular measures. The sixth category, primary findings, captured the main outcomes of the stated study aims. Findings unrelated to the relationship between molecular measures and PTSD were excluded. Extracted data are reported in Table 1.
Table 1. Data extracted from studies examining the molecular contribution to PTSD

Note: The systematic review is limited to original research manuscripts published from 1 January 2022 to 31 December 2023. The primary findings column only reports the results of analyses specifically assessing the relationships between PTSD symptomology and molecular measures. Participant age is reported as mean and standard deviation unless otherwise noted.
Abbreviations: 2-AG, 2-arachidonoylglycerol; Aβ40, amyloid β-40; Aβ42, amyloid β-42; AEA, N-arachidonoylethanalomine; STARRS, Study to Assess Risk and Resilience in Servicemembers; CAPS-5, Clinician-Administered PTSD Scale for DSM5; CAPS-IV, Clinician-Administered PTSD Scale for DSM-IV; CRP, C-reactive protein; DHEA, dehydroepiandrosterone; DMP, differentially methylated position; DMR, differentially methylated region; DNHS, Detroit Neighbourhood Health Study; eQTL, expression quantitative trait loci; GFAP, glial fibrillary acid protein; IES-R, Impact of Event Scale – Revised; IL-1β, interleukin-1 β; IL-10, interleukin-10; IL-6, interleukin-6; LASC, Los Angeles Symptoms Checklist; meQTL, methylation quantitative trait loci; MINI, Mini International Neuropsychiatric Interview; ML, machine learning; MRS, Marine Resilience Study; NFL, neurofilament light chain; NHRVS, National Health and Resilience in Veterans Study; NSE, neuron-specific enolase; PCL-5, PTSD Checklist for DSM5; PCL-C, PTSD Checklist civilian version; PCL-IV, PTSD Checklist for DSM-IV; pNfH, phosphorylated neurofilament heavy chain; PPQ, Perinatal PTSD Questionnaire; PRISMO, Prospective Research in Stress-Related Military Operations; PTS, posttraumatic stress; PTSD, posttraumatic stress disorder; PSS-I, PTSD Symptom Scale – Interview; RICE, Rape Impact Cohort Evaluation; SCID, Structured Clinical Interview for DSM; SRIP, Self-Rating Inventory for PTSD; TNFα, tumour necrosis factor α; TRACTS, Translational Research Center for TBI and Stress Disorders; TSCYC, Trauma Symptom Checklist for Young Children.
Results
Study characteristics
Our search yielded 18 studies that examined longitudinal relationships between PTS/PTSD and molecular biology measures with sample sizes ranging from 39 to 1,135 participants. Study groups were primarily comprised of military veterans/personnel (n = 4), individuals hospitalised following traumatic injury (n = 4), or women exposed to interpersonal violence/trauma (n = 3) or rape (n = 3). The Clinician-Administered PTSD Scale (nDSM-IV = 5, nDSM-5 = 2), PTSD Checklist (nCivilian version = 3, nDSM-5 = 4, nDSM-IV = 2) and Davidson Trauma Scale (n = 3) were the most commonly used measures with both continuous scores and case–control status used as outcome variables. Genomic investigations were the most frequently employed molecular measure and included gene expression (n = 2), genotype (n = 9) and DNA methylation (n = 7) analyses that spanned candidate and whole-genome approaches. Endocrine (n = 2) and inflammatory marker (n = 2) measures were also represented. Four of the studies were performed in LMICs, of which three were conducted in South Africa. The majority of upper-income country (UIC) studies were conducted in the USA (n = 10). Of the 15 studies that reported on ancestry/ethnicity, White/European/Caucasian and African American/Black ancestry were the sole or most prevalent ancestral grouping in seven studies each.
Molecular signatures of PTSD/PTS
Candidate genotype or marker investigations
Several studies employed candidate gene approaches. A study of mother–child dyads found that, in children carrying the ‘risk’ T allele of rs1360780, a variant found in the stress-related FKBP5 gene, higher maternal PTS scores at baseline predicted more severe total and dissociation-related trauma symptoms in children 9 months later. This relationship remained significant even when trauma exposure in the children was controlled for (Pereira et al., Reference Pereira, Printz, Grasso, Hodgkinson, McCarthy, Wakschlag and Briggs-Gowan2021). Landoni et al. (Reference Landoni, Missaglia, Tavian, Ionio and Di Blasio2022) examined the influence of the serotonin transporter long polymorphic region on the development of perinatal PTSD assessed in the 2 months prior to birth (T1), as well as at 2–3 weeks (T2) and 3–4 months (T3) postnatally. The lower expression SS polymorphism was associated with elevated T2 intrusive and T3 hyperarousal symptoms, and moderated the worsening of intrusive, hyperarousal and avoidance symptoms from T1 to T3, intrusive symptoms between T1 and T2, and avoidance symptoms between T2 and T3. Chen et al. (Reference Chen, Zheng, Shen, Guo, Chen, Lin and Fang2022) found that the AA genotype of rs1800872, a single nucleotide polymorphism (SNP) in the gene encoding the anti-inflammatory cytokine interleukin-10 (IL-10), predicted PTSD symptom severity persistence between 6 and 18 months, as well as higher 18-month PTSD scores, in male but not female students following earthquake exposure.
In a study examining endocannabinoids, deRoon-Cassini et al. (Reference deRoon-Cassini, Bergner, Chesney, Schumann, Lee, Brasel and Hillard2022) found that male and ancestry minority (predominantly African) participants admitted to hospital following traumatic injury who were homozygous for the A allele of the FAAH rs324420 SNP had higher baseline serum levels of N-arachidonoylethanalomine compared to C allele carriers. Elevated N-arachidonoylethanalomine positively predicted 6- to 8-month follow-up PTSD status across participants and avoidance and negative alterations of cognition and mood in female participants. Baseline circulating levels of 2-arachidonoylglycerol predicted follow-up reexperiencing, arousal and negative alterations to cognition and mood symptoms in ancestry minority participants. Endocannabinoid signalling is stress responsive and involved in a broad range of biological processes, such as neurotransmission, synaptic plasticity, inflammation, and learning and memory, which may be relevant to PTSD (deRoon-Cassini et al., Reference deRoon-Cassini, Bergner, Chesney, Schumann, Lee, Brasel and Hillard2022; Gorzkiewicz and Szemraj, Reference Gorzkiewicz and Szemraj2018). Vuong et al. sought to identify whether the anti-inflammatory cytokine, adiponectin, may play a role in PTS symptom severity in women following rape exposure. Of the eight SNPs in ADIPOQ, the gene encoding adiponectin, that were investigated, only rs444174 was associated with symptom severity at 3 and 6 months though this relationship did not remain significant when models were adjusted for covariates (Vuong et al., Reference Vuong, Hemmings, Mhlongo, Chirwa, Lombard, Peer, Abrahams and Seedat2022). In a study that included the same rape-exposed participants, as well as unexposed controls, lower baseline serum levels of adiponectin were associated with increased risk of probable PTSD at 6 months across participants. However, this relationship did not hold true when analyses were limited to women exposed to rape (Vuong et al., Reference Vuong, Mhlongo, Chirwa, Lombard, Peer, Hemmings, Abrahams and Seedat2022).
Polygenic risk score analyses
Tamman et al. (Reference Tamman, Wendt, Pathak, Krystal, Southwick, Sippel, Gelernter, Polimanti and Pietrzak2022) found that a higher polygenic risk score, an aggregate score reflecting genetic predisposition liability, predicted increased risk for an incident-positive PTSD screen at 2, 4 or 7 years in military veterans with an insecure attachment style. Of 30 genetic loci significantly associated with PTSD, 13 loci conferred risk or resilience based on patterns of environmental features (trauma burden, age, sex, social and structural support, and combat status). The strongest effect was seen for the rs4702 SNP in the gene encoding FURIN, which has been linked to brain-derived neurotrophic factor and matrix metalloproteinase signalling and plays a role in synaptic plasticity, including in relation to fear and environmental adversity. Risk score gene sets showed enrichment for immune function, specifically biological processes relevant to mast cells. Drug repositioning based on gene ontology highlighted doxylamine, an antihistamine and antimuscarinic that suppresses immune system activation and regulates sleep/wake cycles, as a therapy for investigation.
Epigenetic investigations
Several studies examined epigenetic mechanisms, that is, environmentally sensitive structural changes to DNA conformation that can influence gene expression (Aristizabal et al., Reference Aristizabal, Anreiter, Halldorsdottir, Odgers, McDade, Goldenberg, Mostafavi, Kobor, Binder, Sokolowski and O’Donnell2019). Hawn et al. (Reference Hawn, Neale, Wolf, Zhao, Pierce, Fein‐Schaffer, Milberg, McGlinchey, Logue and Miller2022) examined whether methylation in AIM2, a gene previously linked to inflammation, was associated with PTSD symptom severity and mediated the relationship between PTSD symptomology and markers of inflammation and neuropathology in a cohort of military veterans. The analyses indicated both time- and outcome-dependent effects of methylation. Lower methylation at the cg10636246 site mediated the baseline inverse association between PTSD symptom severity and the neuronal damage marker neurofilament light chain. However, lower cg10636246 site methylation mediated a positive association between symptom severity and proinflammatory (interleukin-6 and tumour necrosis factor alpha) as well as anti-inflammatory cytokines (IL-10) at baseline, as well as an inverse association between symptom severity and IL-10 2 years later. Drawing on measures of social adversity, prior psychopathology and methylation in sites linked to the glucocorticoid receptor response network, Wani et al. (Reference Wani, Aiello, Kim, Xue, Martin, Ratanatharathorn, Qu, Koenen, Galea, Wildman and Uddin2021) used machine learning (ML) to prospectively predict the risk of high PTSD symptom severity in an adult cohort. Though prior PTS symptoms accounted for 88% of the variance, CpG sites made up more than half of the top 150 model features and implicated stress response and inflammatory mechanisms in PTSD risk. Notably, methylation at cg20509117 in IL-6 was among the 79 sites identified. In a study investing intergenerational effects of intimate partner violence-associated PTSD, Cordero et al. (Reference Cordero, Stenz, Moser, Serpa, Paoloni-Giacobino and Schechter2022) aimed to assess correlations between maternal and infant site- and region-level glucocorticoid receptor (NR3C1) methylation profiles and to determine whether maternal methylation profiles predicted offspring internalising and externalising behaviours measured 4 years later. The average methylation of 13 glucocorticoid receptor promoter region sites was significantly correlated between mothers and their infants only in the context of PTSD, that is, not in control mother-infant dyads. A lower maternal methylation profile predicted increased externalising behaviour measured when children were of school-going age. These findings suggest that epigenetic signatures related to stress responding may contribute to the intergenerational effects of PTSD on mental health by biologically embedding adverse outcomes. In a cohort of military personnel with pre- and post-deployment molecular and clinical data, methylation at 4 sites and in 88 differentially methylated regions (DMRs) was associated with baseline PTS symptom severity, whilst longitudinal analysis identified 15 deployment-associated DMRs (Katrinli et al., Reference Katrinli, Maihofer, Wani, Pfeiffer, Ketema, Ratanatharathorn, Baker, Boks, Geuze, Kessler, Risbrough, Rutten, Stein, Ursano, Vermetten, Logue, Nievergelt, Smith and Uddin2022). The two DMRs predicting both baseline and longitudinal PTS symptom severity were located near genes encoding OTUD5 and ELF4, which are involved in inflammatory and oxidative stress processes. In a study investigating epigenetic signatures of PTSD following sexual assault, Nöthling et al. (Reference Nöthling, Abrahams, Toikumo, Suderman, Mhlongo, Lombard, Seedat and Hemmings2021) identified that baseline methylation at an intergenic site near the SLC16A9 gene was higher in rape-exposed women who met criteria for PTSD 3 months later. Two of the 34 PTSD-associated DMRs were located in genes (BRSK2 and ADCYAP1) previously implicated in mood or trauma-related disorders. BRSK2 is involved in neurotransmission and is highly expressed in hippocampus, a brain region fundamental to learning and memory, while the protein encoded by ADCYAP1 plays a key role in stress response regulation. Targeted methylation analyses of regions in BRSK2 and ADCYAP1 were performed in a replication sample, and the longitudinal associations between PTSD symptom and methylation profiles at baseline, 3 and 6 months in both the discovery and replication samples were assessed. Of these follow-up analyses, only an association between changes from baseline to 3 months in PTSD symptom severity and methylation at a BRSK2 site remained significant after controlling for covariates.
Methylation data can be used to generate epigenetic clock estimates of biological ageing, that is, wear and tear that occurs over and above that due to chronological age (Horvath, Reference Horvath2013). Accelerated biological ageing according to DNAm GrimAge, a clock sensitive to age-related morbidity and mortality, in combat veterans was associated with PTSD status at baseline in both discovery and validation cohorts, as well as PTSD symptom severity at baseline when the cohorts were combined. In a subset of discovery cohort participants who completed assessments 3 years later, change in PTSD symptom severity was positively correlated with change in age acceleration, with this relationship primarily driven by symptoms in the hyperarousal cluster. The contribution of immune mechanisms to advanced age acceleration is uncertain given that this biological ageing metric was not associated with circulating levels of five cytokines but was significantly associated with increased immunosenescence based on CD8 and CD28 T lymphocyte proportions (Yang et al., Reference Yang, Gwyneth, Verhoeven, Gautam, Reus, Kang, Flory, Amara, Hood, Doyle, Yehuda, Marmar, Jett, Hammamieh, Mellon and Wolkowitz2021). A study of paramedicine students also found evidence for a relationship between PTSD symptomology and GrimAge estimates with both baseline and one-year GrimAge estimates positively associated with PTSD symptom severity at 1 year (Mehta et al., Reference Mehta, Bruenig, Pierce, Sathyanarayanan, Stringfellow, Miller, Mullens and Shakespeare-Finch2022). Baseline epigenetic age acceleration, a metric derived from the Horvarth algorithm, was positively associated with cross-sectional and longitudinal PTSD symptom severity, whilst baseline, but not follow-up symptom severity, predicted 1-year epigenetic age acceleration.
Studies employing multimodal data
Four studies utilised multilevel data. Schultebraucks et al. (Reference Schultebraucks, Sijbrandij, Galatzer-Levy, Mouthaan, Olff and van Zuiden2021) applied ML models to data routinely collected on emergency room admission and in the 2 days thereafter (endocrine, trauma phenotype, demographic, vital signs, pharmacotherapy, and injury and trauma characteristics) to identify prognostic features for PTSD. The model identified that acute sympathetic nervous system activity, cortisol levels and opiate medication prescription, in addition to age, prior trauma experience and perceived impact of these events, perceived life threat, and amnesia predicted longitudinal (1-, 3-, 6- and 12-month) self-reported symptom severity and 12-month PTSD diagnosis. Three measures of thyroid function were among the 15 strongest predictors, suggesting a role for the hypothalamic–pituitary–thyroid axis. A particular strength of this study was its use of routinely collected data, which provides a higher potential clinical utility. Lori et al. (Reference Lori, Schultebraucks, Galatzer-Levy, Daskalakis, Katrinli, Smith, Myers, Richholt, Huentelman, Guffanti, Wuchty, Gould, Harvey, Nemeroff, Jovanovic, Gerasimov, Maples-Keller, Stevens, Michopoulos, Rothbaum, Wingo and Ressler2021) found that baseline expression levels of GRIN3B, which encodes an NMDA glutamate receptor subunit, in blood were significantly associated with PTS scores at 1, 3, 6 and 12 months and showed a dose-dependent positive relationship with symptom trajectory (chronic vs. remitting vs. resilient) in individuals admitted to hospital emergency units. The authors examined whether methylation and expression quantitative trait loci, that is, loci where methylation or genetic variants, respectively, influence gene expression, could explain the observed relationship between GRIN3B expression and risk for chronic PTSD. The analysis yielded no significant methylation quantitative trait loci, but four SNPs were identified as GRIN3B expression quantitative trait loci with the minor allele of the rs10401454 SNP, which is associated with reduced expression, also associated with resilience to PTSD in an independent study cohort. GRIN3B encodes a subunit of the NMDA glutamate receptor, which may be upregulated in response to stress, and thus implicates glutamatergic processes, such as fear conditioning in the development of PTS symptoms. Furthermore, leukocyte expression of NMDA receptors suggests a tangential link to immune mechanisms in PTSD via GRIN3B effects on NMDAR activity. The study drew on the Genotype-Tissue Expression database to confirm that rs10401454 acts as an expression quantitative trait locus in whole blood, cerebellum and basal ganglia tissue and that minor allele homozygosity is associated with lower GRIN3B expression in cortex and frontal cortex. This indicates a degree of cross-tissue coherence and provides more confidence in inferring brain expression levels based on peripheral tissue measures. Wuchty et al. (Reference Wuchty, Myers, Ramirez-Restrepo, Huentelman, Richolt, Gould, Harvey, Michopolous, Steven, Wingo, Lori, Maples-Keller, Rothbaum, Jovanovic, Rothbaum, Ressler and Nemeroff2021) collected multiomic data from participants immediately and 6 months after trauma exposure. Based on the rationale that dynamic gene expression profiles provide valuable biological insights and that transcriptomic signatures could act as an intermediate phenotype, their study integrated genotype, transcriptomic and gene expression data. Using directed network analysis to draw causal inferences, they identified 13 genes that may drive dysregulated gene expression underlying PTSD development. Key processes supported by these genes are related to threat processing and responding, and fear-related memory acquisition, consolidation, and extinction. Morris et al. (Reference Morris, Sanchez-Sáez, Bailey, Hellman, Williams, Schumacher and Rao2022) used multilevel modelling and ML to examine demographic, cognitive, clinical and biological (diurnal and laboratory stressor-elicited cortisol and alpha-amylase levels, as well as heart rate and hair cortisol) data to predict PTSD development at 1-, 3- and 6-month timepoints in young women exposed to interpersonal violence in the 3 months prior to study initiation. These factors were not associated with baseline or longitudinal PTS symptom severity using multilevel modelling but did improve ML model accuracy, suggesting that the stress response and acute sympathetic nervous system activity play an indirect role in the development of PTSD.
Quality appraisal
All studies adequately described exposure and/or outcome measures, and most assessed potential distorting influences and clearly delineated control or comparison groups where appropriate (Table 2). Limitations were noted in the categories related to the description of the sample and reporting on data. Several studies had relatively modest sample sizes and most did not report on study power. Given the cost of conducting molecular-level investigations, especially those that draw on omics measures, this is not unexpected but does raise doubts as to whether the sample can be considered representative of the source population. The number of potential covariates or confounding influences assessed varied widely across studies. Imputation was frequently employed to address missing data, though several studies did not specifically address the presence/absence of missing data or how this was handled.
Table 2. Quality assessment of included studies according to the systematic appraisal of quality in observational research tool

Abbreviations: 2-AG, 2-arachidonoylglycerol; AEA, N-arachidonoylethanalomine; CIs, confidence intervals; DNHS, Detroit Neighbourhood Health Study; eQTL, expression quantitative trait loci; IL-10, interleukin-10; MAPS, Multidimensional Assessment of Preschoolers; meQTL, methylation quantitative trait loci; ML, machine learning; MRS, Marine Resilience Study; PRISMO, Prospective Research in Stress-Related Military Operations; PTSD, posttraumatic stress disorder; RICE, Rape Impact Cohort Evaluation; SNP, single nucleotide polymorphism; STARRS, Study to Assess Risk and Resilience in Servicemembers; TRACTS, Translational Research Center for TBI and Stress Disorders.
Discussion
Key findings
Molecular profiles
Several key themes emerge from the study findings. First, converging lines of evidence support the role of inflammatory/immune (Wani et al., Reference Wani, Aiello, Kim, Xue, Martin, Ratanatharathorn, Qu, Koenen, Galea, Wildman and Uddin2021; Chen et al., Reference Chen, Zheng, Shen, Guo, Chen, Lin and Fang2022; Hawn et al., Reference Hawn, Neale, Wolf, Zhao, Pierce, Fein‐Schaffer, Milberg, McGlinchey, Logue and Miller2022; Katrinli et al., Reference Katrinli, Maihofer, Wani, Pfeiffer, Ketema, Ratanatharathorn, Baker, Boks, Geuze, Kessler, Risbrough, Rutten, Stein, Ursano, Vermetten, Logue, Nievergelt, Smith and Uddin2022; Tamman et al., Reference Tamman, Wendt, Pathak, Krystal, Southwick, Sippel, Gelernter, Polimanti and Pietrzak2022), stress response (Carleial et al., Reference Carleial, Nätt, Unternährer, Elbert, Robjant, Wilker, Vukojevic, Kolassa, Zeller and Koebach2021; Pereira et al., Reference Pereira, Printz, Grasso, Hodgkinson, McCarthy, Wakschlag and Briggs-Gowan2021; Schultebraucks et al., Reference Schultebraucks, Sijbrandij, Galatzer-Levy, Mouthaan, Olff and van Zuiden2021; Wani et al., Reference Wani, Aiello, Kim, Xue, Martin, Ratanatharathorn, Qu, Koenen, Galea, Wildman and Uddin2021; Cordero et al., Reference Cordero, Stenz, Moser, Serpa, Paoloni-Giacobino and Schechter2022; Morris et al., Reference Morris, Sanchez-Sáez, Bailey, Hellman, Williams, Schumacher and Rao2022) and learning and memory processes (Carleial et al., Reference Carleial, Nätt, Unternährer, Elbert, Robjant, Wilker, Vukojevic, Kolassa, Zeller and Koebach2021; Lori et al., Reference Lori, Schultebraucks, Galatzer-Levy, Daskalakis, Katrinli, Smith, Myers, Richholt, Huentelman, Guffanti, Wuchty, Gould, Harvey, Nemeroff, Jovanovic, Gerasimov, Maples-Keller, Stevens, Michopoulos, Rothbaum, Wingo and Ressler2021; Wuchty et al., Reference Wuchty, Myers, Ramirez-Restrepo, Huentelman, Richolt, Gould, Harvey, Michopolous, Steven, Wingo, Lori, Maples-Keller, Rothbaum, Jovanovic, Rothbaum, Ressler and Nemeroff2021; Tamman et al., Reference Tamman, Wendt, Pathak, Krystal, Southwick, Sippel, Gelernter, Polimanti and Pietrzak2022) in PTSD pathophysiology with exploratory hypothesis-generating approaches utilising whole-genome transcription, epigenetic and genotype data showing enrichment for these mechanisms. Inflammatory processes may contribute to PTSD symptomology by affecting cognition, stress responding, and the structure and function of brain regions involved in affect, and learning and memory (Kim et al., Reference Kim, Lee and Yoon2020; Sumner et al., Reference Sumner, Nishimi, Koenen, Roberts and Kubzansky2020). Two studies directly investigated inflammatory mechanisms. Though the functional effects of the IL-10 SNP rs1800872 investigated by Chen et al. (Reference Chen, Zheng, Shen, Guo, Chen, Lin and Fang2022) is unknown, Hawn et al. (Reference Hawn, Neale, Wolf, Zhao, Pierce, Fein‐Schaffer, Milberg, McGlinchey, Logue and Miller2022) found evidence that a more proinflammatory environment may contribute to PTSD with higher circulating levels of proinflammatory cytokines and lower levels of an anti-inflammatory cytokine predicting worse PTSD symptom severity at baseline and 2 years, respectively. Inflammation is also reciprocally related to the activity of the hypothalamus-pituitary–adrenal axis, which is activated upon threat perception, coordinates the stress response, and may affect cognition and mood (Leistner and Menke, Reference Leistner and Menke2020). The studies by Pereira et al. (Reference Pereira, Printz, Grasso, Hodgkinson, McCarthy, Wakschlag and Briggs-Gowan2021) and Cordero et al. (Reference Cordero, Stenz, Moser, Serpa, Paoloni-Giacobino and Schechter2022) directly assessed components of the stress response system, namely FKBP5 rs1360780 genotype and NR3C1 methylation, respectively. Their findings suggest that the sensitivity and number of glucocorticoid receptors play a role in the pathophysiology of PTSD. This, in addition to findings that including cortisol levels improves predictive performance of ML models (Schultebraucks et al., Reference Schultebraucks, Sijbrandij, Galatzer-Levy, Mouthaan, Olff and van Zuiden2021; Morris et al., Reference Morris, Sanchez-Sáez, Bailey, Hellman, Williams, Schumacher and Rao2022), is in keeping with a substantial body of evidence indicating that dysregulation of the stress response is a key component of PTSD pathophysiology (Fischer et al., Reference Fischer, Schumacher, Knaevelsrud, Ehlert and Schumacher2021). Inflammation and stress responding can accelerate biological ageing and thus underlie the findings of increased epigenetic clock age estimates in PTSD (Wolf and Morrison, Reference Wolf and Morrison2017; Mehta et al., Reference Mehta, Bruenig, Pierce, Sathyanarayanan, Stringfellow, Miller, Mullens and Shakespeare-Finch2022). Identification of processes related to synaptic plasticity, and learning and memory may be explained by previous finding that individuals with PTSD fail to restrict fear responses to the environment, context or cues that elicited the trauma. Instead, overgeneralisation of fearful memories to non-specific targets, combined with reduced fear extinction, can produce the symptoms of hyperarousal and intrusion characteristic of PTSD (Ressler et al., Reference Ressler, Berretta, Bolshakov, Rosso, Meloni, Rauch and Carlezon2022).
The studies included in this participant provide strong evidence for biological processes in PTS. However, the capacity for these biological correlates to predict relative risk or symptom trajectory is not clear. Though genotype investigations are beneficial in that they provide a static measure, PTSD is a polygenic disorder and the contribution of individual variants is likely small (Nievergelt et al., Reference Nievergelt, Maihofer, Klengel, Atkinson, Chen, Choi, Coleman, Dalvie, Duncan, Gelernter, Levey, Logue, Polimanti, Provost, Ratanatharathorn, Stein, Torres, Aiello, Almli, Amstadter, Andersen, Andreassen, Arbisi, Ashley-Koch, Austin, Avdibegovic, Babić, Bækvad-Hansen, Baker, Beckham, Bierut, Bisson, Boks, Bolger, Børglum, Bradley, Brashear, Breen, Bryant, Bustamante, Bybjerg-Grauholm, Calabrese, Caldas-de-Almeida, Dale, Daly, Daskalakis, Deckert, Delahanty, Dennis, Disner, Domschke, Dzubur-Kulenovic, Erbes, Evans, Farrer, Feeny, Flory, Forbes, Franz, Galea, Garrett, Gelaye, Geuze, Gillespie, Uka, Gordon, Guffanti, Hammamieh, Harnal, Hauser, Heath, Hemmings, Hougaard, Jakovljevic, Jett, Johnson, Jones, Jovanovic, Qin, Junglen, Karstoft, Kaufman, Kessler, Khan, Kimbrel, King, Koen, Kranzler, Kremen, Lawford, Lebois, Lewis, Linnstaedt, Lori, Lugonja, Luykx, Lyons, Maples-Keller, Marmar, Martin, Martin, Maurer, Mavissakalian, McFarlane, McGlinchey, McLaughlin, McLean, McLeay, Mehta, Milberg, Miller, Morey, Morris, Mors, Mortensen, Neale, Nelson, Nordentoft, Norman, O’Donnell, Orcutt, Panizzon, Peters, Peterson, Peverill, Pietrzak, Polusny, Rice, Ripke, Risbrough, Roberts, Rothbaum, Rothbaum, Roy-Byrne, Ruggiero, Rung, Rutten, Saccone, Sanchez, Schijven, Seedat, Seligowski, Seng, Sheerin, Silove, Smith, Smoller, Sponheim, Stein, Stevens, Sumner, Teicher, Thompson, Trapido, Uddin, Ursano, van den Heuvel, Van Hooff, Vermetten, Vinkers, Voisey, Wang, Wang, Werge, Williams, Williamson, Winternitz, Wolf, Wolf, Wolff, Yehuda, Young, Young, Zhao, Zoellner, Liberzon, Ressler, Haas and Koenen2019). Therefore, the results of targeted studies, such as those by Chen et al. (Reference Chen, Zheng, Shen, Guo, Chen, Lin and Fang2022), Pereira et al. (Reference Pereira, Printz, Grasso, Hodgkinson, McCarthy, Wakschlag and Briggs-Gowan2021) and Landoni et al. (Reference Landoni, Missaglia, Tavian, Ionio and Di Blasio2022), which identified roles for specific variants in PTSD risk, may not have sufficient discriminative capacity for clinical use. Polygenic risk scores, as generated in Tamman et al. (Reference Tamman, Wendt, Pathak, Krystal, Southwick, Sippel, Gelernter, Polimanti and Pietrzak2022), can explain more of the variance in outcome but do not necessarily perform well across populations of different ancestry. This limitation to their use in global contexts is exacerbated by the underrepresentation of diverse ancestries in psychiatric genetics research (Nievergelt et al., Reference Nievergelt, Maihofer, Klengel, Atkinson, Chen, Choi, Coleman, Dalvie, Duncan, Gelernter, Levey, Logue, Polimanti, Provost, Ratanatharathorn, Stein, Torres, Aiello, Almli, Amstadter, Andersen, Andreassen, Arbisi, Ashley-Koch, Austin, Avdibegovic, Babić, Bækvad-Hansen, Baker, Beckham, Bierut, Bisson, Boks, Bolger, Børglum, Bradley, Brashear, Breen, Bryant, Bustamante, Bybjerg-Grauholm, Calabrese, Caldas-de-Almeida, Dale, Daly, Daskalakis, Deckert, Delahanty, Dennis, Disner, Domschke, Dzubur-Kulenovic, Erbes, Evans, Farrer, Feeny, Flory, Forbes, Franz, Galea, Garrett, Gelaye, Geuze, Gillespie, Uka, Gordon, Guffanti, Hammamieh, Harnal, Hauser, Heath, Hemmings, Hougaard, Jakovljevic, Jett, Johnson, Jones, Jovanovic, Qin, Junglen, Karstoft, Kaufman, Kessler, Khan, Kimbrel, King, Koen, Kranzler, Kremen, Lawford, Lebois, Lewis, Linnstaedt, Lori, Lugonja, Luykx, Lyons, Maples-Keller, Marmar, Martin, Martin, Maurer, Mavissakalian, McFarlane, McGlinchey, McLaughlin, McLean, McLeay, Mehta, Milberg, Miller, Morey, Morris, Mors, Mortensen, Neale, Nelson, Nordentoft, Norman, O’Donnell, Orcutt, Panizzon, Peters, Peterson, Peverill, Pietrzak, Polusny, Rice, Ripke, Risbrough, Roberts, Rothbaum, Rothbaum, Roy-Byrne, Ruggiero, Rung, Rutten, Saccone, Sanchez, Schijven, Seedat, Seligowski, Seng, Sheerin, Silove, Smith, Smoller, Sponheim, Stein, Stevens, Sumner, Teicher, Thompson, Trapido, Uddin, Ursano, van den Heuvel, Van Hooff, Vermetten, Vinkers, Voisey, Wang, Wang, Werge, Williams, Williamson, Winternitz, Wolf, Wolf, Wolff, Yehuda, Young, Young, Zhao, Zoellner, Liberzon, Ressler, Haas and Koenen2019). Other genomic approaches also have limitations. Studies utilising epigenetic and transcriptomic data report relative associations that is, the analyses are based on comparisons within study groups and do not provide set cut-off values that could be used to stratify individuals according to risk. Methylation and transcriptomic measures can differ according to tissue type and cellular composition, the latter of which most, but not all, of the studies reviewed stated that they controlled for. Studies also indicated that the influence of biological mechanisms may depend on environmental as well as demographic factors such as sex and ancestry (Chen et al., Reference Chen, Zheng, Shen, Guo, Chen, Lin and Fang2022; deRoon-Cassini et al., Reference deRoon-Cassini, Bergner, Chesney, Schumann, Lee, Brasel and Hillard2022; Tamman et al., Reference Tamman, Wendt, Pathak, Krystal, Southwick, Sippel, Gelernter, Polimanti and Pietrzak2022). A further limitation in identifying biological correlates with potential clinical utility is that PTSD can have diverse presentations with recent studies indicating the presence of multiple typologies such as threat reactivity, low-symptom, high-symptom and dysphoric classes (Campbell et al., Reference Campbell, Trachik, Goldberg and Simpson2020; Bucich et al., Reference Bucich, Steel and Berle2022). This phenotypic complexity may be mirrored in the biological processes conferring risk for specific symptom clusters, as was seen in the studies by Landoni et al. (Reference Landoni, Missaglia, Tavian, Ionio and Di Blasio2022), deRoon-Cassini et al. (Reference deRoon-Cassini, Bergner, Chesney, Schumann, Lee, Brasel and Hillard2022) and Yang et al. (Reference Yang, Gwyneth, Verhoeven, Gautam, Reus, Kang, Flory, Amara, Hood, Doyle, Yehuda, Marmar, Jett, Hammamieh, Mellon and Wolkowitz2021). Consequently, it is also possible that existing and valid relationships between biological correlates and symptom typologies were not discerned in studies that used only PTSD status and/or total symptom scores. Based on these limitations, biological correlate research at present seems better able to inform on underlying mechanisms than to stratify individuals according to risk and target preventative measures and interventions.
Study contexts
Selected studies were mainly conducted in UICs and comprised predominantly Caucasian ancestry participants, although those recruiting patients admitted to hospital emergency units and military veterans/personnel reflected a more diverse ancestry profile. Despite being home to 84.5% of the global population in 2021 (World Bank, n.d.), less than one-quarter of the selected studies were conducted in LMICs. These investigations reflect a more constrained participant demographic profile (Black adult females or Chinese male and female adolescents), range of trauma experiences (rape or earthquake exposure) and number of molecular techniques employed (candidate gene, inflammatory marker or DNA methylation) than UIC studies. None of the studies generating multimodal data or using ML or multilevel modelling were performed in LMICs. Representation within LMIC studies was also unequal with three of four publications based on a single South African cohort study examining outcomes of rape exposure in female participants that identified as Black African. Our search criteria did not yield any studies conducted in South American or South or South-East Asian populations. Furthermore, all but one study recruited adult participants. This is problematic given that the age structure in LMICs is skewed towards younger age categories and that trauma exposure occurring during sensitive windows of neurodevelopment can have substantial effects on neurobiology and mental health risk across the lifespan (Herringa, Reference Herringa2017). The treatment gap is higher in LMICs with treatment-seeking rates in these countries only half that reported in UICs, and established risk factors for PTSD, including social disadvantage, younger age, lower household income, unemployment and lower levels of education, are observed at disproportionately higher rates in LMICs (Koenen et al., Reference Koenen, Ratanatharathorn, Ng, McLaughlin, Bromet, Stein, Karam, Ruscio, Benjet, Scott, Atwoli, Petukhova, Lim, Aguilar-Gaxiola, Al-Hamzawi, Alonso, Bunting, Ciutan, de Girolamo, Degenhardt, Gureje, Haro, Huang, Kawakami, Lee, Navarro-Mateu, Pennell, Piazza, Sampson, Ten Have, Torres, Viana, Williams, Xavier and Kessler2017). These LMIC vs. UIC disparities undermine efforts to address the global burden of PTSD. Understanding the interaction of environmental and molecular neurobiological determinants in the genesis of PTSD is key to the discovery of new and effective treatments, which has been highlighted as a particularly pressing need in LMICs. However, the potential for biological mechanisms to inform preventative and therapeutic measures will only be realised if the number and scope of studies more closely align with the global burden of trauma and PTSD.
Study approaches
Several studies integrated multimodal data which can provide comprehensive and deep insights into psychiatric outcomes (Wuchty et al., Reference Wuchty, Myers, Ramirez-Restrepo, Huentelman, Richolt, Gould, Harvey, Michopolous, Steven, Wingo, Lori, Maples-Keller, Rothbaum, Jovanovic, Rothbaum, Ressler and Nemeroff2021). Studies made use of sophisticated analyses, including ML and multilevel modelling. The former does not test specific hypotheses but rather learns which combination of features best predicts outcomes and, by including variable interrelationships, can account for indirect contributions to outcomes. In contrast, multilevel modelling assesses how variables are related and can thus provide a more interpretable, but potentially less accurate, model (Morris et al., Reference Morris, Sanchez-Sáez, Bailey, Hellman, Williams, Schumacher and Rao2022). Unfortunately, the cost of collecting multilevel data limits study replication and sample size, the latter of which reduces the likelihood of adequately representing the source population or conducting sex-, age- or ancestry-stratified analyses. Therefore, it is important that LMICs are included in psychiatric genetic and other consortia of multi-country investigators and studies. Despite relatively small sample sizes, omics studies can increasingly draw on available public and consortia databases to interpret and gain functional insights into data. Examples include functional enrichment for biological processes and information on tissue-specific gene expression. The latter is particularly useful in neuroscience, where the inaccessibility of brain tissue necessitates inferences about neural mechanisms based on peripheral tissues. Though the results obtained from multiomics studies are informative, their clinical utility is low insofar as they rely on data that is not routinely collected from or available for individuals seeking post-trauma care (Schultebraucks et al., Reference Schultebraucks, Sijbrandij, Galatzer-Levy, Mouthaan, Olff and van Zuiden2021).
Limitations
This review has several limitations. We selected a narrow publication timeframe and limited our selection to longitudinal studies reporting results in English. This excludes insights offered by studies published in other languages and by cross-sectional investigations, and may have limited the representation of selected studies. Though the number of publications fitting our search terms is increasing year-on-year (Figure 2), it is not possible to discern whether this is true for both UICs and LMICs, or to draw inferences about trends in methodological approaches. The narrow timeframe, as well as the impact of COVID-19, could also have exaggerated the fault line between published research in UICs and LMICs. Our focus on molecular-level aetiology only reflects a subset of biological mechanisms and ignores findings from neuroimaging and psychophysiological studies. We did not report on animal model investigations, which have the potential to unravel causal mechanisms, as opposed to the correlative associations identified in human studies. We also did not conduct a meta-analysis of study findings. Finally, we did not consider the full range of trauma-associated adverse outcomes or the contribution of molecular profiles to cross-disorder risk.
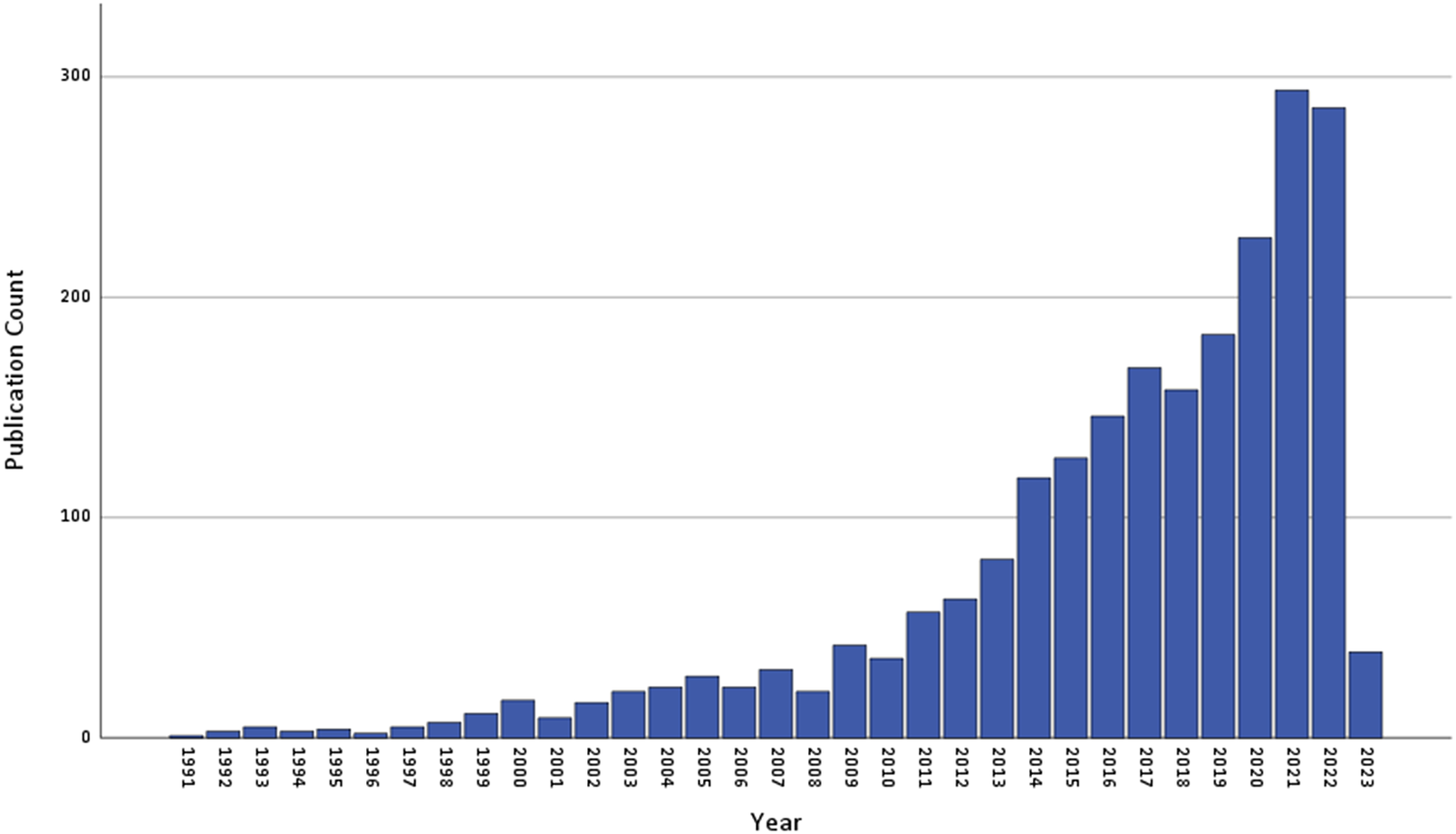
Figure 2. Annual count of number of publications retrieved from PubMed using the systematic review search string. The results are based on the following query: (("post-traumatic stress" (All Fields)) OR ("posttraumatic stress" (All Fields)) OR (PTSD (All Fields))) AND ((neurobiolog* (All Fields)) OR (genom* (All Fields)) OR (DNA (All Fields)) OR ("stress hormone" (All Fields))) AND (English(Language)) NOT (animal). The search was limited to studies published up to 31 December 2022.
Conclusions
The potential benefits of biological insights into PTSD are compelling with recent research identifying inflammatory, stress response, and learning and memory processes in PTSD pathophysiology. The sophistication and scope of molecular techniques and data analytic approaches are rapidly expanding. However, greater representation of LMICs in research is required for biological insights to reduce the global burden of PTSD and associated adverse effects on health and wellbeing.
Open peer review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/gmh.2023.53.
Data availability statement
A systematic review of published literature was performed. No original research data were generated.
Author contribution
J.S.W.: Conceptualisation, investigation, data curation, writing – original draft, writing – review and editing. M.P.: Investigation, data curation, writing – original draft, writing – review and editing. M.C.G.: Investigation, data curation, writing – original draft, writing – review and editing. L.L.H.: Conceptualisation, writing – review and editing. E.K.: Writing – review and editing. S.S.: Conceptualisation, writing – review and editing.
Financial support
Research reported in this publication was partly supported by the South African Medical Research Council. The work by JSW was made possible through funding by the South African Medical Research Council through its Division of Research Capacity Development under the Early Investigators Programme from funding received from the South African National Treasury. The content hereof is the sole responsibility of the authors and does not necessarily represent the official view of the SAMRC. MCG was supported by a Career Development Award from the U.S. National Institute of Mental Health (K01MH129572).
Competing interest
The authors declare no competing interests exist.
Ethics standard
All authors declare that they have adhered to the publishing ethics of Global Mental Health.








Comments
Dear editor,
On behalf of my co-authors, I am pleased to submit the following invited review "Advances in the molecular neurobiology of posttraumatic stress disorder from global contexts: A systematic review of longitudinal studies” to Global Mental Health.
Trauma exposure is a globally prevalent phenomenon that can exert profound and lasting effects on health and wellbeing. Nevertheless, only a subset of exposed individuals will go on to develop posttraumatic stress disorder, which is characterised by intrusive thoughts, avoidance behaviours, hypervigilance and negative alterations in cognition and mood. A substantial body of research indicates that biological mechanisms play an important role in determining risk and resilience, and advances in molecular technologies and data analytic procedures now provide unprecedented insights into the molecular aetiology underlying PTSD. These findings have the potential to reduce the burden of posttraumatic stress disorder on global mental health. Specifically, identification of biological correlates of posttraumatic stress disorder could be used to stratify trauma-exposed individuals according to risk, target prevention and intervention initiatives to those at highest risk, identify biological targets for potential therapeutic intervention and track treatment response.
In this systematic review, we aimed to summarise the findings of studies investigating the longitudinal relationships between molecular-level measures and posttraumatic stress disorder status or symptom severity. We chose to focus on studies published in 2021 and 2022 so as to provide a state-of-the-art overview. The 28 studies that met our inclusion criteria recruited participants with diverse experiences of trauma, including military conflict, interpersonal violence, sexual assault, traumatic injury and natural disasters. We provide an overview of the study results and find strong evidence for inflammation/immune, stress response and learning and memory processes as key biological signatures in the pathophysiology of posttraumatic stress disorder. We have highlighted the methodological advances in the field. Our manuscript also contains an embedded discussion of the nature and form of the literature in upper- compared to lower- and middle-income countries. The majority of studies included in the systematic review, and especially those that used multilevel data, were conducted in upper-income countries, which indicates a mismatch between trauma research and the global burden of trauma exposure and associated outcomes.
We believe that our manuscript is well suited to the aims and scope of the journal, as it provides an accessible yet comprehensive overview of recent research into the molecular neurobiology of posttraumatic stress disorder, a globally prevalent psychiatric disorder. We would like to thank you for granting us permission to exceed the 4000 word limit, which has allowed us to provide a more detailed overview of a substantial body of research. We would also like to confirm that this manuscript has not been previously published and is not under consideration by another journal.
We hope that you consider our manuscript favourably.
Sincerely,
Jacqueline Womersley