Introduction
Dorylaimida Pearse, Reference Pearse1942 is one of the most diverse orders in terms of number of species within the phylum Nematoda (Jairajpuri & Ahmad Reference Jairajpuri and Ahmad1992; Andrassy Reference Andrássy2009). The family Longidoridae Thorne, Reference Thorne1935 (Thorne Reference Thorne1935), inside Dorylaimida, includes obligate plant ectoparasitic species and is one of the most economically important nematode groups in agriculture. The importance of this group of nematodes lies not only in their polyphagy and cosmopolitan distribution but also their status as vectors of plant viruses that cause significant damage to a wide range of agricultural crops (Coomans Reference Coomans1996; Taylor & Brown Reference Taylor and Brown1997; Macfarlane Reference Macfarlane2003; Decraemer & Robbins Reference Decraemer and Robbins2007; Decraemer & Geraert Reference Decraemer, Geaert, Perry and Moens2013; Archidona-Yuste et al. Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillo2016a, Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillob, Reference Archidona-Yuste, Wiegand, Castillo and Navas-Cortés2019a). The family Longidoridae includes more than 500 species (Coomans et al. Reference Coomans, Huys, Heyns and Luc2001; Decraemer & Robbins Reference Decraemer and Robbins2007), where the needle nematodes of the genus Longidorus Micoletzky, Reference Micoletzky1922 are one of the most diverse genera of this family. This genus includes a number of long to very long body (2–12 mm) specimens with long stylet (80–260 μm). They are a polyphagous species of many plants including various agricultural crops, and they cause damage by direct feeding on root cells as well as by transmitting nepoviruses (nepoviruses are icosahedral, with a bipartite positive stranded RNA genome, wherein each RNA encodes as a single polyprotein). The genus Longidorus is a diverse group with more than 177 nominal species (Gutiérrez-Gutiérrez et al. Reference Gutiérrez-Gutiérrez, Teixeira Santos, Inácio, Eisenback and Mota2020; Clavero-Camacho et al. Reference Clavero-Camacho, Liébanas, Escuer, Cantalapiedra-Navarrete, Archidona-Yuste, Castillo and Palomares-Rius2021). Only 11 species (6.9%) (L. apulus Lamberti & Bleve-Zacheo, Reference Lamberti and Bleve-Zacheo1977; L. arthensis Brown et al., Reference Brown, Grunder, Hooper, Klingler and Kunz1994; L. attenuatus Hooper, Reference Hooper1961; L. caespiticola Hooper, Reference Hooper1961; L. diadecturus Eveleigh & Allen, Reference Eveleigh and Allen1982, L. elongatus (de Man, Reference De Man1876) Thorne & Swanger, Reference Thorne and Swanger1936; L. fasciatus Roca & Lamberti, Reference Roca and Lamberti1981; L. leptocephalus Hooper, Reference Hooper1961; L. macrosoma Hooper, Reference Hooper1961; L. martini Merny, Reference Merny1966, and L. profundorum Hooper, Reference Hooper1965) have been reported as virus vector transmitting seven nepoviruses (artichoke Italian latent virus, cherry rosette disease virus, tomato black ring virus, raspberry ringspot virus, Arabis mosaic virus, peach rosette mosaic virus, and mulberry ringspot virus) (Brown et al. Reference Brown, Lamberti, Taylor and Trudgill1988; Taylor & Brown Reference Taylor and Brown1997; Decraemer & Robbins Reference Decraemer and Robbins2007). These nematodes spend their entire life cycle in the rhizosphere, using their needle stylet to feed on the apical root cells, inducing galls in the tips and arresting root growth (Taylor & Brown Reference Taylor and Brown1997; Palomares-Rius et al. Reference Palomares-Rius, Escobar, Cabrera, Vovlas and Castillo2017). The morphological convergence and the existence of cryptic species in this genus make the accurate identification of species considerably more difficult (De Luca et al. Reference De Luca, Reyes, Grunder, Kunz, Agostinelli, De Giorgi and Lamberti2004; Gutiérrez-Gutiérrez et al. Reference Gutiérrez-Gutiérrez, Cantalapiedra-Navarrete, Montes-Borrego, Palomares-Rius and Castillo2013; Archidona-Yuste et al. Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillo2016b, Reference Archidona-Yuste, Cantalapiedra-Navarrete, Castillo and Palomares-Rius2019b). Consequently, morphological taxonomy could lead to underestimation of the diversity in the genus Longidorus as reported in other genera of plant-parasitic nematodes (Palomares-Rius et al. Reference Palomares-Rius, Cantalapiedra-Navarrete and Castillo2014; Archidona-Yuste et al. Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillo2016a, Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillob, Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castilloc; Janssen et al. Reference Janssen, Karssen, Orlando, Subbotin and Bert2017). Therefore, accurate identification of Longidorus species is essential in establishing appropriate control measures and control strategies for preventing the spread of these nematodes. To date, 27 Longidorus species have been reported from Iran, including: L. aetnaeus Roca et al., Reference Roca, Lamberti, Agostinelli and Vinciguerra1986; L. africanus Merny, Reference Merny1966; L. apulus; L. armeniacae Bakhshi Amrei et al., Reference Bakhshi Amrei, Peneva, Rakhshandehroo and Pedram2022; L. artemisiae Rubtsova et al., Reference Rubtsova, Chizhov and Subbotin1999; L. behshahrensis Bakhshi Amrei et al., Reference Bakhshi Amrei, Peneva, Rakhshandehroo and Pedram2020; L. crassus Thorne, Reference Thorne1974; L. elongatus; L. euonymus Mali & Hooper, Reference Mali and Hooper1974; L. hyrcanus Mobasseri et al., Reference Mobasseri, Pourjam, Farashiani and Pedram2023; L. iranicus Sturhan & Barooti, Reference Sturhan and Barooti1983; L. kheirii Pedram et al., Reference Pedram, Niknam, Robbins, Ye and Karegar2008; L. leptocephalus; L. orientalis Loof, Reference Loof1982; L. paravineacola Ye & Robbins, Reference Ye and Robbins2003; L. perangustus Roshan-Bakhsh et al., Reference Roshan-Bakhsh, Pourjam and Pedram2016; L. persicus Esmaeili et al., Reference Esmaeili, Heydari, Archidona-Yuste, Castillo and Palomares-Rius2017; L. pisi Edward et al., Reference Edward, Misra and Singh1964; L. profundorum; L. protae Lamberti & Bleve-Zacheo, Reference Lamberti and Bleve-Zacheo1977; L. proximus Sturhan & Argo, Reference Sturhan and Argo1983; L. sabalanicus Asgari et al., Reference Asgari, Eskandari, Castillo and Palomares-Rius2022; L. soosanae Pour Ehtesham et al., Reference Pour Ehtesham, Pedram, Atighi and Jahanshahi Afshar2023; L. sturhani Rubtsova et al., Reference Rubtsova, Subbotin, Brown and Moens2001; L. tabrizicus Niknam et al., Reference Niknam, Pedram, Ghahremani Nejad, Ye, Robbins and Tanha Maafi2010, and L. vineacola Sturhan & Weischer, Reference Sturhan and Weischer1964. In a May 2022 survey, a population of an unidentified species of Longidorus was recovered from the rhizosphere of Astragalus sp. naturally growing in the mountains of the Anguran Protected Area, west Mahneshan, Zanjan province. Molecular approaches and phylogenetic studies in combination with morphometric characters are used as a taxonomic standard for species identification and delimitation, which is known as Integrative Taxonomy (Gutiérrez-Gutiérrez et al. Reference Gutiérrez-Gutiérrez, Cantalapiedra-Navarrete, Montes-Borrego, Palomares-Rius and Castillo2013; Peneva et al. Reference Peneva, Lazarova, De Luca and Brown2013; Archidona-Yuste et al. Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillo2016d). Our research aims to characterize this undescribed nematode species based on morphological characters integrated with molecular data and infer the phylogenetic relationships with the other species of genus Longidorus.
Materials and methods
Nematode population sampling, extraction, and morphological identification
About 100 soil samples were collected from the rhizosphere of different plants at a depth of 10–50 cm, in the Zanjan province, north-western Iran. Specimens of an unidentified Longidorus sp. nov. were obtained from the rhizosphere of Astragalus sp. in Zanjan province. Nematodes were extracted using the tray method (Whitehead & Hemming Reference Whitehead and Hemming1965), the magnesium sulphate (MgSO4) centrifugal flotation method (Coolen Reference Coolen, Lamberti and Taylor1979), and a modification of Cobb’s decanting and sieving method (Flegg Reference Flegg1967). Nematodes were handpicked under a stereomicroscope, killed by adding hot FPG (4:1:1, formaldehyde: propionic acid: glycerin) solution, transferred to anhydrous glycerine according to De Grisse (Reference De Grisse1969), and mounted on permanent glass slides to allow handling and observation. Morphometric values and photomicrographs were taken using a Dino-Eye digital eyepiece camera (Model AM7023, bundled with DinoCapture 2.0 software; AnMo Electronics Corporation, New Taipei City, Taiwan) attached to a Leitz Dialux 22 light microscope. Line drawings were first made using a drawing tube, then re-drawn and prepared for publication using CorelDRAW software version 16 (Corel Corp, Canada). Morphological comparisons were performed using the polytomous identification keys for the identification of Longidorus species (Chen et al. Reference Chen, Hooper, Loof and Xu1997; Loof & Chen Reference Loof and Chen1999) and with the descriptions of all other characterized species up to the present. The position of pharyngeal gland nuclei was calculated according to Loof & Coomans (Reference Loof and Coomans1972), and the juvenile developmental stages were identified according to Robbins et al. (Reference Robbins, Brown, Halbrendt and Vrain1995). All measurements were recorded in micrometres (μm), except for body length in millimetres (mm) and ratios. Ratios are defined in Jairajpuri & Ahmad (Reference Jairajpuri and Ahmad1992).
Molecular characterization
For the molecular phylogenetic studies, four live nematode specimens (two females and two juveniles) were selected. Each specimen was transferred to an Eppendorf tube containing 10 μl ddH2O, 8 μl lysis buffer (125 mM KCl, 25 mM Tris–Cl pH 8.3, 3.75 mM MgCl2, 2.5 mM DTT, 1.125% Tween 20, 0.025% gelatine), and 2 μl proteinase K (600 μg/ml), and crushed for 2 min with a micro-homogeniser (Subbotin et al. Reference Subbotin, Halford, Warry and Perry2000). The tubes were frozen at -80 °C (15 min), then incubated at 65 °C (1 h) and at 95 °C (10 min), consecutively. After centrifugation (1 min, 16,000 × g), 4 μl of extracted DNA were added to the polymerase chain reaction (PCR) mixture in a 0.2 ml Eppendorf tube containing: 20 μl 2 × Master mix (Amplicon, Odense, Denmark), 2 μl of each primer (10 pMol/μl), and 12 μl ddH2O, to a final volume of 40 μl. The D2-D3 expansion segments of 28S rRNA were amplified using forward D2A (5′–ACAAGTACCGTGAGGGAAAGTTG–3′) and reverse D3B (5′–TCGGAAGGAACCAGCTACTA–3′) primers (Nunn Reference Nunn1992). The ITS1 region was amplified using forward primer 18S (5′–TTGATTACGTCCCTGCCCTTT–3′) (Vrain et al. Reference Vrain, Wakarchuk, Levesque and Hamilton1992) and reverse primer rDNA1 5.8S (5′–ACGAGCCGAGTGATCCACCG–3′) (Cherry et al. Reference Cherry, Szalanski, Todd and Powers1997). PCR reactions were carried out in a DNA thermal cycler (Hybaid, Ashford, Middlesex, UK), and the amplification program was set as follows: initial denaturation at 94 °C for 10 min; followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C (LSU), 55 °C (ITS1) for 30 s, extension at 72 °C for 1 min; and finally, the elongation step at 72 °C for 6 min. The amplified PCR products were purified using ExoSAP-IT (Affimetrix, USB products), quantified using a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA), and used for direct sequencing in both directions using the primers referred to above. The resulting products were purified and run on a DNA multicapillary sequencer (Model 3130XL genetic analyser; Applied Biosystems, Foster City, CA, USA), using the BigDye Terminator Sequencing Kit v.3.1 (Applied Biosystems, Foster City, CA, USA) at the Stab Vida sequencing facilities (Caparica, Portugal). The newly obtained sequences were submitted to the GenBank database under the accession numbers OR509844-OR509847 for D2-D3 expansion segments of 28S rRNA and OR509848-OR509851 for ITS1 region.
Phylogenetic analyses
The newly obtained sequences of L. zanjanensis sp. nov. (D2-D3 expansion segments of 28S rRNA, and ITS1 rRNA) and other sequences of different Longidorus spp. from GenBank were used for phylogenetic analyses. ITS1 rRNA did not have enough similarity with other sequences deposited in the GenBank, and for this reason, sequence similarity comparisons were only made with the closest phylogenetically related species. Outgroup taxa for each dataset were chosen following previously published studies (He et al. Reference He, Subbotin, Rubtsova, Lamberti, Brown and Moens2005; Holterman et al. Reference Holterman, van der Wurff, van den Elsen, van Megen, Bongers, Holovachov, Bakker and Helder2006; Palomares-Rius et al. Reference Palomares-Rius, Subbotin, Landa, Vovlas and Castillo2008; Gutiérrez-Gutiérrez et al. Reference Gutiérrez-Gutiérrez, Cantalapiedra-Navarrete, Montes-Borrego, Palomares-Rius and Castillo2013; Archidona-Yuste et al. Reference Archidona-Yuste, Cantalapiedra-Navarrete, Castillo and Palomares-Rius2019b; Cai et al. Reference Cai, Archidona-Yuste, Cantalapiedra-Navarrete, Palomares-Rius and Castillo2020a, Reference Cai, Prior, Lawson, Cantalapiedra-Navarrete, Palomares-Rius, Castillo and Archidona-Yusteb). Multiple sequence alignments for each gene were made using the FFT-NS-2 algorithm of MAFFT V.7.450 (Katoh et al. Reference Katoh, Rozewicki and Yamada2019). Sequence alignments were visualized using BioEdit (Hall Reference Hall1999) and manually edited and trimmed of poorly aligned positions using a light filtering strategy (up to 20% of alignment positions), which has little impact on tree accuracy and may save some computation time, as suggested by Tan et al. (Reference Tan, Muffato, Ledergerber, Herrero, Goldman, Gil and Dessimoz2015). Phylogenetic analyses of the sequence datasets were based on Bayesian inference (BI) using MrBayes 3.1.2 (Ronquist & Huelsenbeck Reference Ronquist and Huelsenbeck2003). The best-fit model of DNA evolution was obtained using JModelTest V.2.1.7 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012) with the Akaike Information Criterion (AIC). The best-fit model, base frequency, proportion of invariable sites, and gamma distribution shape parameters and substitution rates in the AIC were then used in MrBayes for the phylogenetic analyses. The general time-reversible model with invariable sites and a gamma-shaped distribution (GTR + I + G) for the D2-D3 segments of 28S rRNA and the general time-reversible model and a gamma-shaped distribution (GTR + G) for ITS1 rRNA were run with four chains for 4 × 106 generations, respectively. The Markov chains were sampled at intervals of 100 generations. Two runs were conducted for each analysis. After discarding burn-in samples of 30% and evaluating convergence, the remaining samples were retained for in-depth analyses. The topologies were used to generate a 50% majority-rule consensus tree. Posterior probabilities (PP) were given on appropriate clades. Trees from all analyses were visualised using FigTree software version 1.4.4 (Rambaut Reference Rambaut2018).
Results and Discussion
The integration of nematode morphology with the morphometric analysis and molecular data using ribosomal sequences allowed us to describe herein a new species of the genus as L. zanjanensis sp. nov.
Longidorus zanjanensis sp. nov.
Zoobank: urn:lsid:zoobank.org:act:7584860D-A994-495B-A5CD-5AE62DBA0AD6
Description

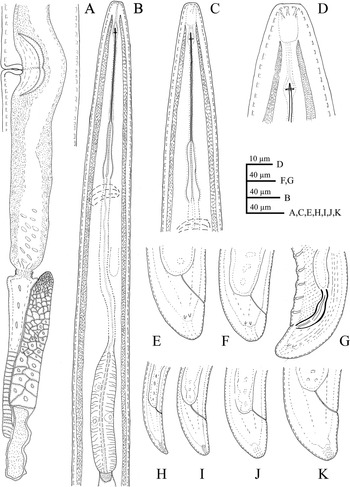
Figure 1. Line drawings of Longidorus zanjanensis sp. nov. A: Female reproductive system. B: Female anterior region. C: Female anterior region. D: Anterior end showing amphidial fovea. E, F: Female tail. G: Male posterior body region. H: J1 tail. I: J2 tail. J: J3 tail. K: J4 tail.
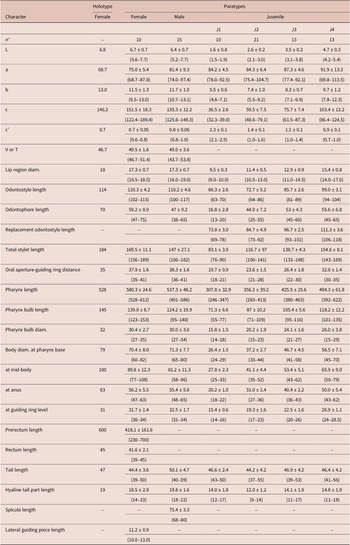
Table 1. Morphometrics of Longidorus zanjanensis sp. nov. from Zanjan, Iran. All measurements are in μm (except L, in mm) and in the form: mean ± standard deviation (range)
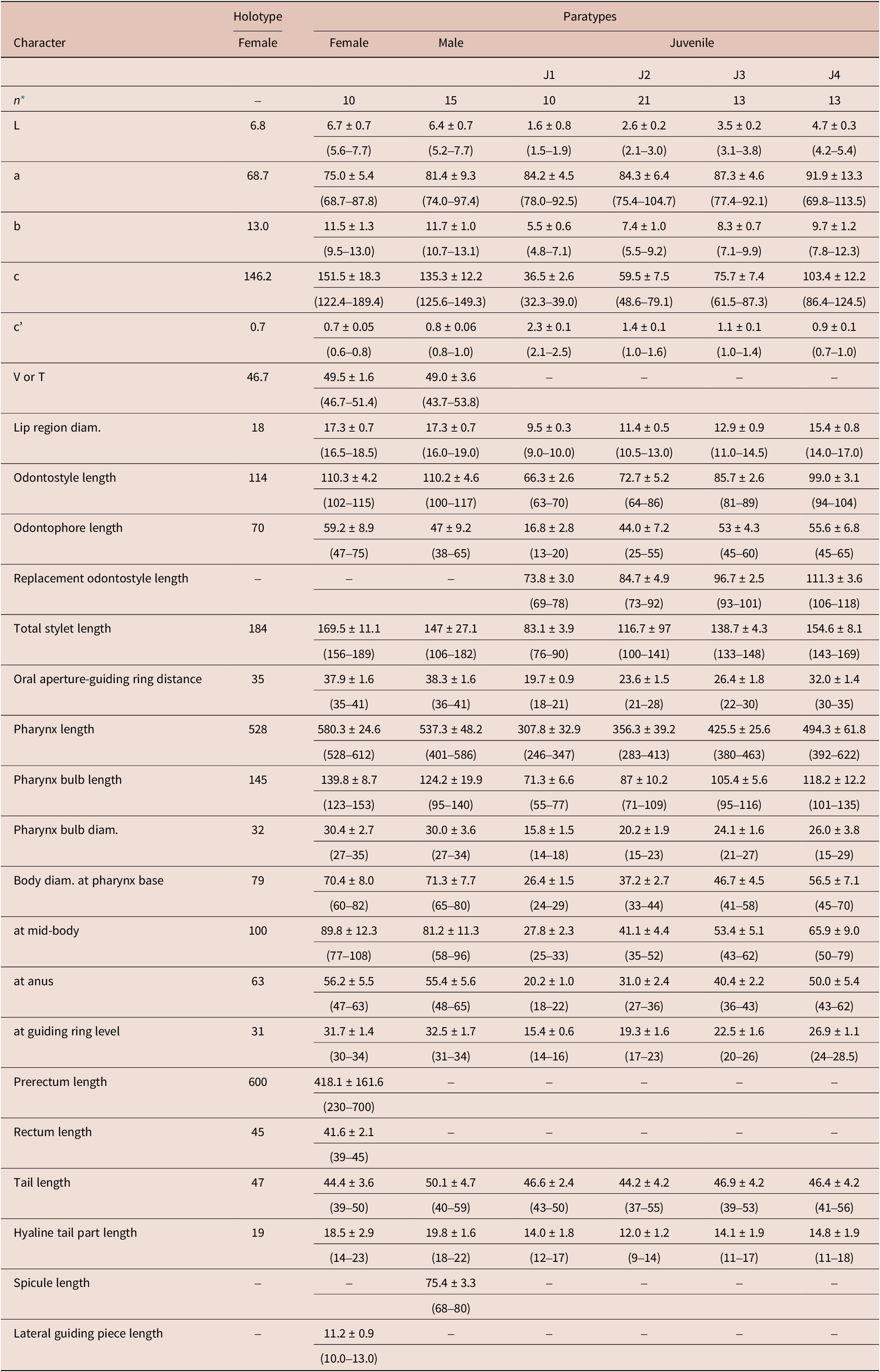
* Abbreviations: n = number of specimens on which measurements are based; L = overall body length; a = body length/greatest body diameter; b = body length/distance from anterior end to pharyngo-intestinal junction; c = body length/tail length; c’ = tail length/tail diameter at anus or cloaca; V = distance from body anterior end to vulva expressed as percentage (%) of the body length; T = distance from cloacal aperture to anterior end of testis expressed as percentage (%) of the body length.
Female. Body ventrally bent varying from J to G shape when heat-relaxed. Cuticle appearing smooth under light microscope; its thickness varies over the body, 4–5 μm at guiding ring level, to 3–4 μm at mid-body, 7–8 μm at anterior lip of anus, and 14–23 μm at tail end (the hyaline part of tail tip), and marked by very fine superficial transverse striae mainly in tail region. Lateral chord 23–30% of corresponding body diameter. Lip region anteriorly flattened, continuous with the adjacent body (Figures 1, 2). Amphidial fovea is pouch-like without lobes at base. Stylet guiding ring located at ca. two times lip region diameter from anterior end. Odontostyle long and narrow, approximately 1.9 times as long as odontophore (Figures 1, 2). Nerve ring surrounding the slender portion of the pharynx posterior to the odontophore base, located at 247–280 μm from anterior end. Pharynx dorylaimoid, anterior slender part flexible, posteriorly expanding to a muscular terminal bulb occupying about 24.1 ± 1.7 (20.7–27.4)% of the total pharynx (neck region).The dorsal gland nucleus (DN) smaller, at 26.1–36.4%, and the two ventrosublateral nuclei (S1N) at about the same level and at 51.0–61.5% of the pharyngeal bulb length (location of glands nuclei according to Loof & Coomans (Reference Loof and Coomans1972)). Cardia conoid to rounded, 5.0–7.0 μm long. Intestine with prerectum. The reproductive system didelphic–amphidelphic, with both branches almost equally developed, each branch 500–1100 μm long, with reflexed ovaries highly variable in length, anterior ovary (173–324 μm long), and posterior ovary (157–390 μm long). Oviducts slightly longer than ovaries. Uterus bipartite, quite variable in length, anterior uteri (230–276) μm long, and posterior uteri (208–286) μm long; sphincter well developed, between uterus and oviduct. Sperm commonly found in the uteri. Vagina 50–74 μm long or ca. 66% of corresponding body width; pars distalis 11–16 μm long, pars proximalis vaginae measuring 20–34 μm long; vulva a transverse slit. Prerectum variable in length, 3.9–11.4 times longer than anal body width and rectum simple, 0.8–0.1 times as long as tail length. Tail bearing two caudal pores, conoid, convex dorsally, and ventrally almost straight or slightly concave with rounded terminus.

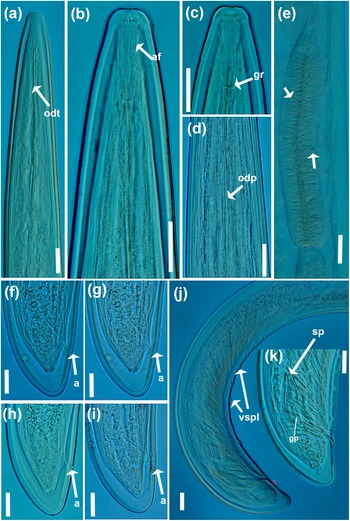
Figure 2. Light micrographs of Longidorus zanjanensis sp. nov. (a–d) female anterior body regions showing odontostyle, odontophore, amphidial fovea, and guiding ring (arrowed); (e) detail of basal bulb showing dorsal gland and ventrosublateral nuclei (arrowed); (f-i) female tail; (j, k) male tail with details of spicules, guiding pieces of gubernaculum and ventromedian supplements (arrowed). Abbreviations: a = anus; af = amphidial fovea; gr = guiding ring; gp = guiding pieces of gubernaculum; odt = odontostyle; odp = odontophore; sp = spicule; vspl = ventromedian supplement. (Scale bars: 20 μm).
Male. Common (about 60% of the population) and functional. Similar to females in general morphology, except for the reproductive system and posterior end more ventrally curved Male genital reproductive system diorchic. Spicules arcuate, robust, about 1.5 times longer than tail length, lateral guiding piece more or less straight. Adanal supplements paired, preceded anteriorly by a row of 8–14 irregularly spaced ventromedian supplements. Tail bluntly conoid, dorsally convex and ventrally concave, terminus widely rounded, with distinct radial lines in hyaline region. Tail length almost equivalent to cloacal body width.
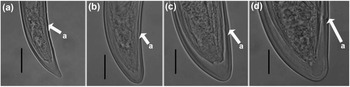
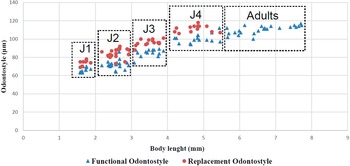
Juveniles. Morphologically similar to adults in most respects except for size and development of reproductive system. All juvenile developmental stages were detected and distinguished by relative lengths of body and functional and replacement odontostyle (Figure 4). J1 characterized by a conoid tail, dorso-ventrally curved with rounded terminus, and slight depression at hyaline region level, with a c´ ratio average of 2.3, odontostyle length ca. 66.3 μm, and shorter distance from anterior end to stylet guiding-ring than that in adult stages. For the rest of the juvenile stages (J2, J3, J4), the replacement odontostyle were located at some distance posterior to the odontophore base and morphology of tail were similar to females (bluntly conoid with a rounded terminus, dorsally convex and ventrally almost straight or slightly concave), becoming stouter after each moult (Figure 3).

Figure 3. Light micrographs of Longidorus zanjanensis sp. nov. (a–d) tails of J1, J2, J3, and J4. (Scale bars: a–d = 20 μm).
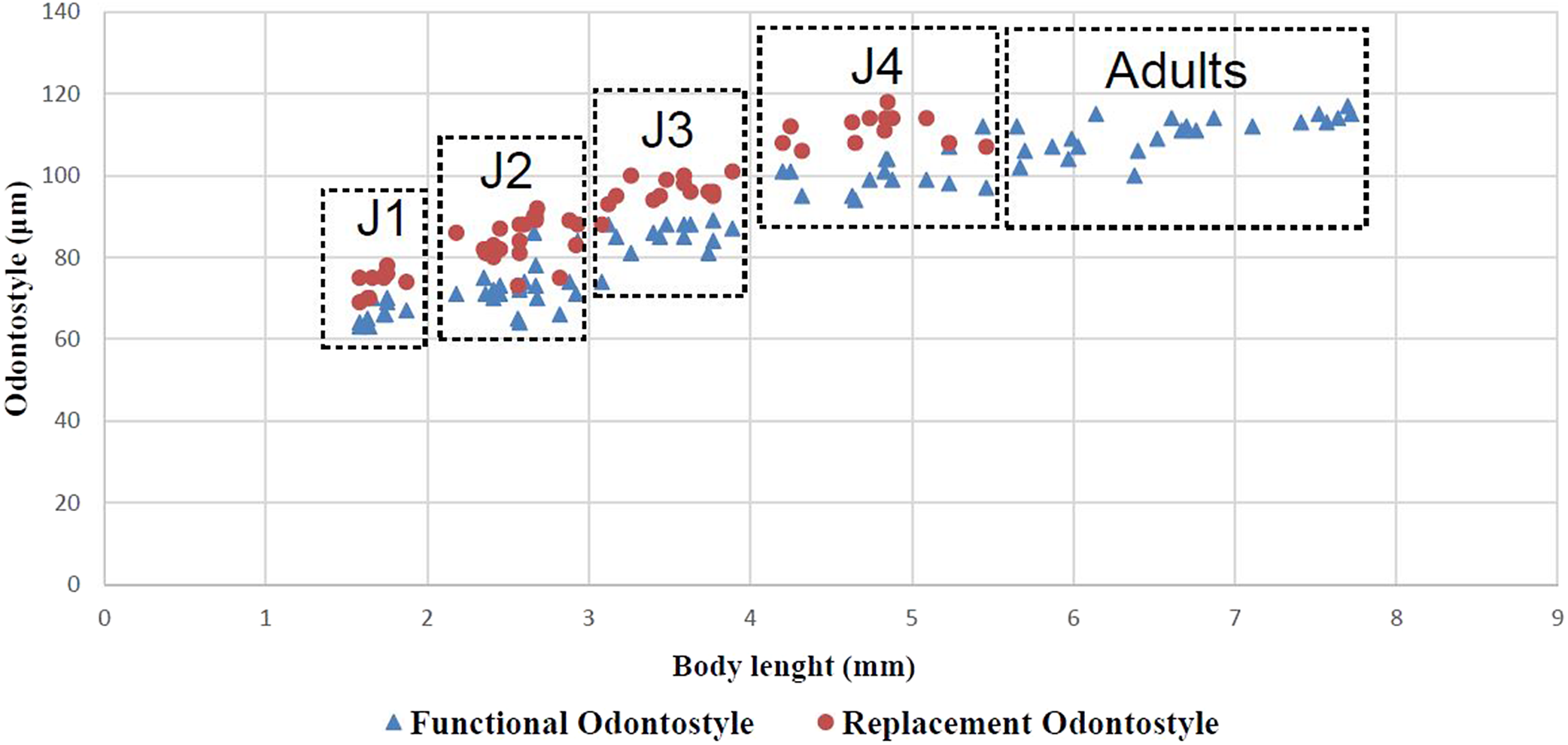
Figure 4. Relationship between body length and functional and replacement odontostyle length in all developmental juvenile life stages and mature adults of Longidorus zanjanensis sp. nov.
Diagnosis and relationships
Longidorus zanjanensis sp. nov. is characterised by a thick (a < 88) and long body (5.6–7.7 mm); lip region 16.5–18.5 μm wide and continuous with body contour; amphidial fovea pouch shaped, not bilobed, and extending about 1/2 part of oral aperture-guiding ring distance; relatively long odontostyle (102–115 μm); guiding ring located at 35.0–41.0 μm from anterior end; vulva located at 46.7–51.4% of body length; female tail short and bluntly conoid (39.0–50.0 μm long, c = 122.4–189.4, c’ = 0.6–0.8), with two pairs of caudal pores. Males with long spicules (68–80 μm) and 8–14 ventromedian supplements. Four developmental juvenile stages were identified, with the first stage juvenile with conoid tail (c’ = 2.1–2.5). According to the polytomous key by Chen et al. (Reference Chen, Hooper, Loof and Xu1997), supplement by Loof & Chen (Reference Loof and Chen1999), and the addition of some characters by Peneva et al. (Reference Peneva, Lazarova, De Luca and Brown2013), codes for the new species are: A4-B3-C3-D3-E1-F34-G12-H1-I2-J1-K6; and specific D2-D3 expansion segments of 28S rRNA, and ITS1 region (GenBank accession numbers: OR509844 for D2-D3 expansion segments of 28S rRNA and OR509848-OR509851 for ITS1 region).
According to the body and odontostyle length, shape of amphidial fovea, distance of guiding ring from anterior body end, lip region and tail shape, a and c’ ratios, and frequency of males, the new species is close to eight known Longidorus species, namely L. apulus; L. armeniacae; L. crassus; L. ferrisi Robbins et al., Reference Robbins, Ye and Pedram2009; L. kheirii; L. pauli Lamberti et al., Reference Lamberti, Molinari, De Luca, Agostinelli and Di Vito1999; L. proximus; and L. soosanae. In addition, L. zanjanensis sp. nov. is closely related molecularly to L. hyrcanus; L. elongatus; and also L. soosanae.
The new species differs from L. apulus by having a different amphidial fovea shape (pouch-like-shaped, not bilobed vs. symmetrically bilobed at base), a higher oral aperture to guiding ring distance (35.0–41.0 vs. 24.0–34.0 μm), lower a ratio (68.7–87.8 vs. 110–154), and longer spicule length (68.0–80.0 vs. 57.0 μm). From L. armeniacae, it differs by a higher oral aperture to guiding ring distance (35.0–41.0 vs. 29.0–35.0 μm), shorter spicule length (68.0–80.0 vs. 80.0–107 μm), and different tail shape in J1 (slender conical, without a digitate or subdigitate terminus vs. convex-conoid to conical, with a distinctly digitate terminus). It differs from L. crassus by having a longer body (5.6–7.7 vs. 5.0–6.0 mm), different amphidial fovea shape (pouch-like, not bilobed vs. symmetrically bilobed at base), wider lip region width (16.5–18.5 vs. 15 μm), higher oral aperture to guiding ring distance (35.0–41.0 vs. 32.5 μm), and lower a ratio (68.7–87.8 vs. 80.0–107 μm).
From L. ferrisi, it differs by a longer body (5.6–7.7 vs. 4.3–5.9 mm), different amphidial fovea shape (pouch-like, not bilobed vs. symmetrically bilobed at base), and longer spicule length (68.0–80.0 vs. 53.0–63 μm). From L. kheirii, it differs by having a smaller body (5.6–7.7 vs. 6.7–9.0 mm), smaller odontostyle and odontophore (102–115 vs. 113–130 μm and 47.0–75.0 vs. 69.0–97.5), narrower lip region width (16.5–18.5 vs. 19.5–23.0 μm), smaller tail (39.0–50.0 vs. 47.0–72.0 μm), and smaller spicule length (68.0–80.0 vs. 85.0 μm). From L. pauli, it differs by a smaller body [average 6.7 (5.6–7.7) vs. average 7.6 (6.5–8.6 mm)], different amphidial fovea shape (pouch-like, not bilobed vs. asymmetrically bilobed at base), wider lip region width (16.5–18.5 vs. 13.9–16.8 μm), lower a ratio (68.7–87.8 vs. 120.3–143.5), higher oral aperture to guiding ring distance (35.0–41.0 vs. 27.2–35.8 μm), longer spicule length (68.0–80.0 vs. 61.0–69 μm), and a lower number of ventromedian supplements in the male tail (8–14 vs. 12–16). From L. proximus, it differs by a lip region shape (continuous vs. expanded, high, separated from the rest of body by a depression), lower a and c ratio (68.7–87.8 vs.104–138 and 122.4–189.4 vs. 165–249), and position of pharyngeal gland nuclei (normal vs. more posterior). It differs from L. soosanae by having an anterior body region shape (uniformly narrowing towards anterior end vs. bottle-shaped), longer odontostyle (102–115 vs. 92.0–103 μm), lower a ratio (68.7–87.8 vs. 79–114), longer tail (39–50 vs. 33–42 μm), and longer spicule length (68.0–80.0 vs. 50.0–64 μm). From L. hyrcanus, it differs by a longer body (5.6–7.7 vs. 5.0–5.8 mm), different shaped lip region (anteriorly flattened, continuous with the adjacent body vs. rounded, continuous with body contour), different amphidial fovea shape (not bilobed vs. asymmetrically bilobed at base), wider lip region width (16.5–18.5 vs. 11.5–14.0 μm), longer spicule length (68.0–80.0 vs. 55.0–68 μm), different tail shape in J1 (slender conical, without a digitate or subdigitate terminus vs. slender, with broadly rounded tail end), and shorter tail of J1 (43.0–50.0 vs. 24.0 μm). Finally, from L. elongatus, the new species differs mainly by having a longer body (5.6–7.7 vs. 4.5–6.4 mm), different amphidial fovea shape (pouch-like, not bilobed vs. asymmetrically bilobed at base), longer odontostyle (102–115 vs. 81.0–102 μm), and higher oral aperture to guiding ring distance (35.0–41.0 vs. 29.0–36.0 μm).
Etymology
The specific epithet refers to the province of Zanjan, north-western Iran where the new species was collected.
Type host and locality
The type population was collected from the rhizosphere of Astragalus sp. naturally growing in mountains of Anguran Protected Area, West Mahneshan, Zanjan province, north-western Iran, coordinates: 36º 51’ 9.5416’’ N; 47º 45’ 1.2196’ ’E; altitude: 1350 m a. s. l.
Type material
Holotype female (slide: 1412-b5), five female paratypes, 11 male paratypes, and 57 juvenile paratypes (slides: 1412-b1-16) were deposited in the nematode collection at the Faculty of Agriculture, University of Zanjan, Zanjan, Iran. Four female paratypes and four male paratypes were deposited in the nematode collection at the Institute for Sustainable Agriculture (IAS), Spanish National Research Council (CSIC), Cordoba, Spain (IAS_L_2023-2_Ir).
The Life Science Idenitifier (LSID) for the publication is: urn:lsid:zoobank.org:pub:E071D8F1- E494-4D1F-9DDF-782D6BDD3CCF.
Molecular characterisation and phylogenetic position of Longidorus zanjanensis sp. nov.
The amplification of D2-D3 segments of 28S rRNA and ITS1 yielded single fragments of ca. 900 bp, and 1100 bp, respectively, based on gel electrophoresis. Four identical sequences were obtained for D2-D3 segments of 28S rRNA with 97.3% (22 nucleotides difference), 96.9% (23 nucleotides difference), and 96.4% (27 nucleotides difference) similarity with L. soosanae (ON122993), L. hyrcanus (OL739253-OL739254), and several sequences of L. elongatus (MN123751), respectively. Four ITS1 region sequences with only one nucleotide difference in one sequence were obtained. Similarly to the D2-D3 region, the ITS1 region is similar to L. soosanae (ON121993-ON121994), L. hyrcanus (OL684817), and several sequences of L. elongatus (AF511417), at 87.0–88.5%, 89.2%, and 85.5%, respectively.
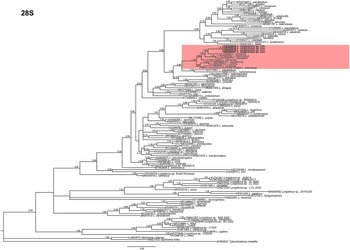
Phylogenetic relationships among Longidorus species inferred from analyses of D2-D3 expansion segments of 28S rRNA and ITS1 sequences using BI are given in Figures 5 and 6, respectively. The D2-D3 expansion segments of the 28S rRNA tree of Longidorus spp. are based on a multiple edited alignment including 121 sequences and 760 total characters, revealing four major clades, three of them highly supported (PP = 1.00) and the other with moderate support (PP = 0.97) (Figure 5). Longidorus zanjanensis sp. nov. (OR509844-OR509847) and L. hyrcanus (OL739254) clustered in a moderately supported clade (PP = 0.96). These two species are related to L. intermedius Kozlowska & Seinhorst, Reference Kozlowska and Seinhorst1979 (AY593058), L. elongatus (AF480078), and L. soosanae (ON122993) in a relatively well-supported clade (PP = 0.98); they are related in another relatively well-supported clade (PP = 0.96) to L. carpathicus Lišková et al., Reference Lišková, Robbins and Brown1997 (AF480072), L. uroshis Krnjaić et al., Reference Krnjaić, Lamberti, Krnjaić, Agostinelli and Radicci2000 (EF538754), L. piceicola Lišková, et al., Reference Lišková, Robbins and Brown1997 (KY086070), and L. artemisiae (KX137849). These species from both clades are related in a highly supported clade (PP = 1.00).

Figure 5. Phylogenetic relationships of Longidorus zanjanensis sp. nov. within the genus Longidorus. Bayesian 50% majority rule consensus tree as inferred from D2 and D3 expansion domains of 28S rRNA sequence alignment under the general time-reversible model of sequence evolution with correction for invariable sites and a gamma-shaped distribution (GTR + I+ G). Posterior probabilities more than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in boldface type, and coloured box indicates clade association of the new species. Scale bar = expected changes per site.

Figure 6. Phylogenetic relationships of Longidorus zanjanensis sp. nov. within the genus Longidorus. Bayesian 50% majority rule consensus tree as inferred from ITS1 region sequence alignment under the GTR + G model. Posterior probabilities more than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in boldface type, and coloured box indicates clade association of the new species. Scale bar = expected changes per site.
For the ITS1 region sequences, the 50% majority rule consensus BI tree of a multiple sequence alignment containing 12 sequences and 928 characters is showed in Figure 6. Longidorus zanjanensis sp. nov. (OR509848-OR509851) clustered with L. soosanae (ON121994) in a low supported clade (PP = 0.83). These two species clustered with L. hyrcanus in a highly supported clade (PP = 0.99). Additionally, L. elongatus (AF511417) is closely related to these three species in a low-supported clade (PP = 0.90) sp. nov. These species are related with L. piceicola (LT669803) and L. intermedius (KT308890) in a highly supported clade (PP = 1.00).
This new species increases the knowledge of the biodiversity of this genus in Iran, including molecular markers for its unequivocal identification. Other species from Iran are closely related to our species (L. soosane and L. hyrcanus), but clearly separated using our integrative taxonomy. This species is clearly described using an integrative taxonomical approach (combination of morphology-morphometry and molecular data). The high diversity of this genus in Iran points to this region as a high diversity location for this group of nematodes.
Acknowledgements
The authors thank Mrs. C. Cantalapiedra-Navarrete (IAS-CSIC) for helping with molecular analyses, and anonymous reviewers and editors for their efforts in reviewing the manuscript and helping improve this study.
Financial support
There are not projects associated with this research.
Competing interest
None.
Ethical standard
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.