INTRODUCTION
Piroplasmoses are tick-borne diseases caused by apicomplexan protistan haemoparasites of the genera Babesia, Theileria and Cytauxzoon that have a great economic, veterinary and medical impact worldwide. While Theileria and Cytauxzoon species infect mainly ungulates and felids, respectively, Babesia spp. infect a wide range of avian and mammalian hosts, including humans (Dobbelaere and Kuenzi, Reference Dobbelaere and Kuenzi2004; Meinkoth and Kocan, Reference Meinkoth and Kocan2005; Schnittger et al. Reference Schnittger, Rodriguez, Florin-Christensen and Morrison2012). Babesiidae are considered the second most common haemoparasites of mammals after trypanosomes (Schnittger et al. Reference Schnittger, Rodriguez, Florin-Christensen and Morrison2012). Many Babesia species of domestic carnivores and ungulates have been reported in genetically close-related wildlife species, causing clinical manifestation when placed in captivity in non-native areas (Penzhorn, Reference Penzhorn2006). Babesiosis is considered a worldwide emerging zoonosis with Babesia microti and Babesia divergens being the most common causes of human babesiosis in the USA and/or Europe (Yabsley and Shock, Reference Yabsley and Shock2013). Wildlife species are reservoir for several Babesia spp. (Yabsley and Shock, Reference Yabsley and Shock2013) but the vectors of many species are still unknown. Theileria annae, which is phylogenetically close to zoonotic B. microti, was first described in a German dog after travelling to Galicia (Zahler et al. Reference Zahler, Rinder, Schein and Gothe2000), a northwestern region of Spain where this species is considered endemic in domestic dogs (Canis lupus familiaris) (Camacho, Reference Camacho2006). Since then, it has been identified in dogs and foxes in North America and Europe (Gimenez et al. Reference Gimenez, Casado, Criado-Fornelio, de Miguel and Dominguez-Penafiel2009; Birkenheuer et al. Reference Birkenheuer, Horney, Bailey, Scott, Sherbert, Catto, Marr, Camacho and Ballman2010; Dezdek et al. Reference Dezdek, Vojta, Curkovic, Lipej, Mihaljevic, Cvetnic and Beck2010; Cardoso et al. Reference Cardoso, Cortes, Reis, Rodrigues, Simoes, Lopes, Vila-Vicosa, Talmi-Frank, Eyal, Solano-Gallego and Baneth2013; Najm et al. Reference Najm, Meyer-Kayser, Hoffmann, Herb, Fensterer, Pfister and Silaghi2014; Miro et al. Reference Miro, Checa, Paparini, Ortega, Gonzalez-Fraga, Gofton, Bartolome, Montoya, Galvez, Mayo and Irwin2015). Phylogenetic studies carried out recently (Baneth et al. Reference Baneth, Florin-Christensen, Cardoso and Schnittger2015), confirmed that T. annae does not belong to the genus Theileria and that it can be clearly distinguished from B. microti infecting rodents, macaques, and humans. Instead, it segregates into a single clade within the group of Babesia that infect carnivores. Since the red fox (Vulpes vulpes) seems to be its natural reservoir, authors proposed to re-name this haemoparasite as Babesia vulpes, replacing all others synonyms previously used such as Babesia ‘Spanish dog’ isolate, B. microti-like or Babesia annae (Baneth et al. Reference Baneth, Florin-Christensen, Cardoso and Schnittger2015). Ixodes hexagonus is suspected to be the tick vector of this agent (Camacho et al. Reference Camacho, Pallas, Gestal, Guitian, Olmeda, Telford and Spielman2003). Furthermore, a new piroplasm related to B. vulpes was found in a European badger (Meles meles) in Spain (Gimenez et al. Reference Gimenez, Casado, Criado-Fornelio, de Miguel and Dominguez-Penafiel2009), but knowledge about this species is still limited.
In the past decades, populations of badgers and red foxes have increased and are adapted to peri-urban and urban environments across Europe (Gloor et al. Reference Gloor, Bontadina, Hegglin, Deplazes and Breitenmoser2001; Judge et al. Reference Judge, Wilson, Macarthur, Delahay and McDonald2014). Therefore, these carnivores represent a possible source of pathogens for domestic animals and humans. This study was carried out to investigate by molecular methods piroplasms infection in wild carnivores, including Canidae, Mustelidae, Felidae and Viverridae species from Northern Spain.
MATERIALS AND METHODS
Study site
The study was carried out in two autonomous regions in northern Spain (Asturias and the Basque Country). Samples were collected from 121 sites located in the Basque Country and 28 from Asturias. The Basque Country (7234 km2) has an Atlantic Climate, with mild to slightly hot summers and mild winters (AEMET, 2011). The temperate climate of the region allows for a varied and abundant wildlife fauna. Regarding wild carnivores, the most abundant species are badger and red fox. Asturias (10 603 km2) has also an Atlantic Climate but precipitations are more abundant and winters are colder than in the Basque Country, with frequent snow falls in the mountainous areas from October till May (AEMET, 2011). Wildlife in this region is very abundant and diverse, including carnivores such as red fox, badger, wolf (Canis lupus) and the endangered brown bear (Ursus arctos). Climate and host abundance do also allow for high burdens of ticks in the environment (Barandika et al. Reference Barandika, Berriatua, Barral, Juste, Anda and García-Pérez2006).
Wildlife sampling
Sample collection was conducted in the frame of a larger study on surveillance of zoonotic infectious diseases in wild animals in both Spanish regions. A total of 241 wild carnivores (169 collected in the Basque Country and 72 in Asturias), belonging to eight Mustelidae, two Canidae, one Viverridae and one Felidae species were processed (Table 1). Red foxes were hunted mostly in winter (65%). The remaining wild species were collected throughout the year after being found dead or moribund. A complete necropsy was performed at the laboratory. Spleen tissue samples were stored at –20 °C before being processed for PCR analysis. Whenever possible, animals were subjected to a detailed external examination and attached ticks were removed, counted and identified using specific taxonomic keys (Manilla, Reference Manilla1998).
Table 1. Distribution of infection as determined by RTi-PCR and tick infestation among wild carnivores
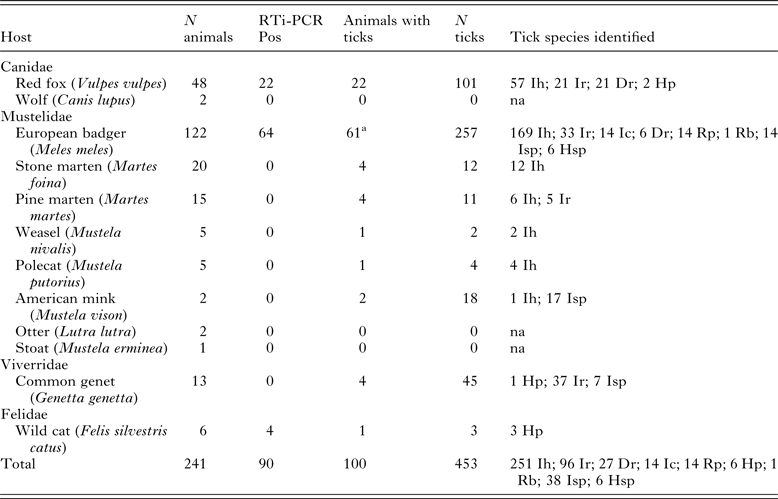
Ih, I. hexagonus; Ir, I. ricinus; Ic, I. canisuga; Dr, D. reticulatus; Rp, R. pusillus; Hp, H. punctata; Rb, R. bursa; Isp, Ixodes sp.; Hsp, Haemaphysalis sp.; na, non-applicable
a Three animals were not subject to external examination.
Molecular analysis
DNA extraction was performed on individual spleen samples using BioSprint 96 DNA kit (Qiagen) with a previous treatment with proteinase K for 3 h. Negative extraction controls were included every ten samples to rule out any possible contamination in the extraction procedure. Presence of piroplasms was firstly determined using a real-time PCR assay (RTi-PCR) that targets the 18S rRNA gene of the genera Babesia and Theileria; the assay also includes an internal amplification control to monitor for possible inhibition. Sequences of primers and probes, as well as details on cycling conditions, sensitivity and specificity were as reported elsewhere (Hurtado et al. Reference Hurtado, Barandika, Oporto, Minguijón, Povedano and García-Pérez2015). Analyses were performed in 20 µL volume reactions using an ABI PRISM 7500 Fast Sequence Detection System (Applied Biosystems). Samples were analysed along with the extraction negative controls and, at least two non-template negative controls (sterile molecular-grade water) per amplification reaction were included.
PCR-positive samples were identified by sequencing analysis of either the V4 region or the nearly complete 18S rRNA gene (one representative for each taxon). Briefly, an about 1600-bp fragment of the 18S rRNA gene was amplified from all positive samples using primers 18STBF (5′-AACCTGGTTGATCCTGCCAGT-3′) and 18STBR (5′-GATCCTTCYGCAGGTTCACC-3) and about 750 nt, that included the V4 region, were sequenced using primer RLB-R2 (5′-CTAAGAATTTCACCTCTGACAGT-3). Larger fragments were sequenced for one representative of each taxon using primers Nbab_1F (5′-AAGCCATGCATGTCTAAGTATAAGCTTTT-3′) and Nbab1_R (5′-CCTCTCCTTCCTTTAAGTGATAAGGTTCAC-3′) or specific primers designed by primer walking for Hepatozoon sp. Sequencing reactions were carried out using the ABI BigDye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) and products were analysed on an ABI 3130 Genetic Analyser (Applied Biosystems, Foster City, CA, USA).
Nucleotide sequence accession numbers
Nucleotide sequence data reported in this paper are available in the GenBank database under the accession numbers: KT223483–86, KU198329–30.
Data analysis
The obtained sequences were identified by comparison with the GenBank database by homology searches made at the National Center for Biotechnology Information (NCBI) using BLASTN. For phylogenetic analyses, a multiple sequence alignment was performed using Mega 6 package (Tamura et al. Reference Tamura, Stecher, Peterson, Filipski and Kumar2013) with an engine based on the MUSCLE algorithm (Edgar, Reference Edgar2004). Selected taxa for the analysis of 18S rRNA gene sequences included the closest BLASTN hits to the sequences described here, along with representatives of B. microti and other Babesia species, including those that infect carnivores. Toxoplasma gondii and Eimeria maxima were included as outgroup to root the tree. The phylogenetic tree was then inferred by using the Maximum Likelihood method based on the Tamura–Nei model (Tamura and Nei, Reference Tamura and Nei1993) and the Gamma distributed with Invariant Sites (G+I) option for variation in rates among sites (model with the lowest Bayesian Information Criterion). Accuracy of inferred topology was assessed via bootstrap analysis (Felsenstein, Reference Felsenstein1985) of 1000 replicates.
Prevalences of piroplasm infection and different piroplasms species in relation to ticks and carnivore species were compared with Chi-square and Fisher exact tests. Statistical analyses were carried out with the SAS statistical package (SAS Institute 9.3, Cary, NC, USA). P values <0.05 were considered statistically significant.
RESULTS
DNA isolated from spleen samples from the 241 carnivores were analysed by real-time PCR, detecting a positive signal in 90 (37.3%) of them. Nine wild carnivore species were negative, though in six of them only five or fewer specimens were analysed. Positive animals included wild cats (Felis silvestris catus) (4/6; 66.7%), badgers (64/122; 52.5%) and red foxes (22/48; 45.8%) (Table 1).
A total of 68 samples were successfully sequenced producing six different sequence types (Table 2); sequencing attempts failed for a further 22 samples (5 foxes and 17 badgers). The four samples from wild cat were identical among themselves and to Cytauxzoon sp. from a Eurasian lynx (Lynx lynx) from Romania (GenBank accession no. KT361080). Close homology (99.7%) was also found to Cytauxzoon sp. from wild cats from Romania (KT361072), an Iberian lynx (Lynx pardinus) from Spain (AY496273) or a domestic cat (F. silvestris catus) from France (EU622908). The 17 amplicons sequenced from red foxes were confirmed as being 100% identical to B. vulpes (AF188001 – Babesia sp. ‘Spanish dog’). Conversely, variability was found in haemoparasites infecting badgers, where four different sequence types of three different genera were identified. In four badgers, a sequence closely related to Cystoisospora spp. was found (99.2% identical to Cystoisospora timoni EU200792 from Suricata suricatta and 99.1% to Cystoisospora belli JX025651). In one of them, Cystoisospora sp. was present as a mixed infection with an Hepatozoon species 99.7% (1635/1640 nt) identical to that found in European pine marten (Martes martes) (EF222257). However, the most abundant haemoparasite among badgers was Babesia, represented by two sequence types referred here as Babesia sp. badger types A and B. Both types only shared 96.7% (1275/1318) similarity between them and about 97–98% homology with their best hits in GenBank, AF188001 (Babesia sp. ‘Spanish dog’) and FJ645725 (Theileria sp. HN2), and were found in 36 and 7 badgers, respectively (Table 2). Phylogenetic analysis of the babesias found in this study confirmed the identity of the piroplasms from red fox as B. vulpes and showed that the two Babesia sp. from badger clearly clustered with Babesia spp. that infect carnivores (Fig. 1).
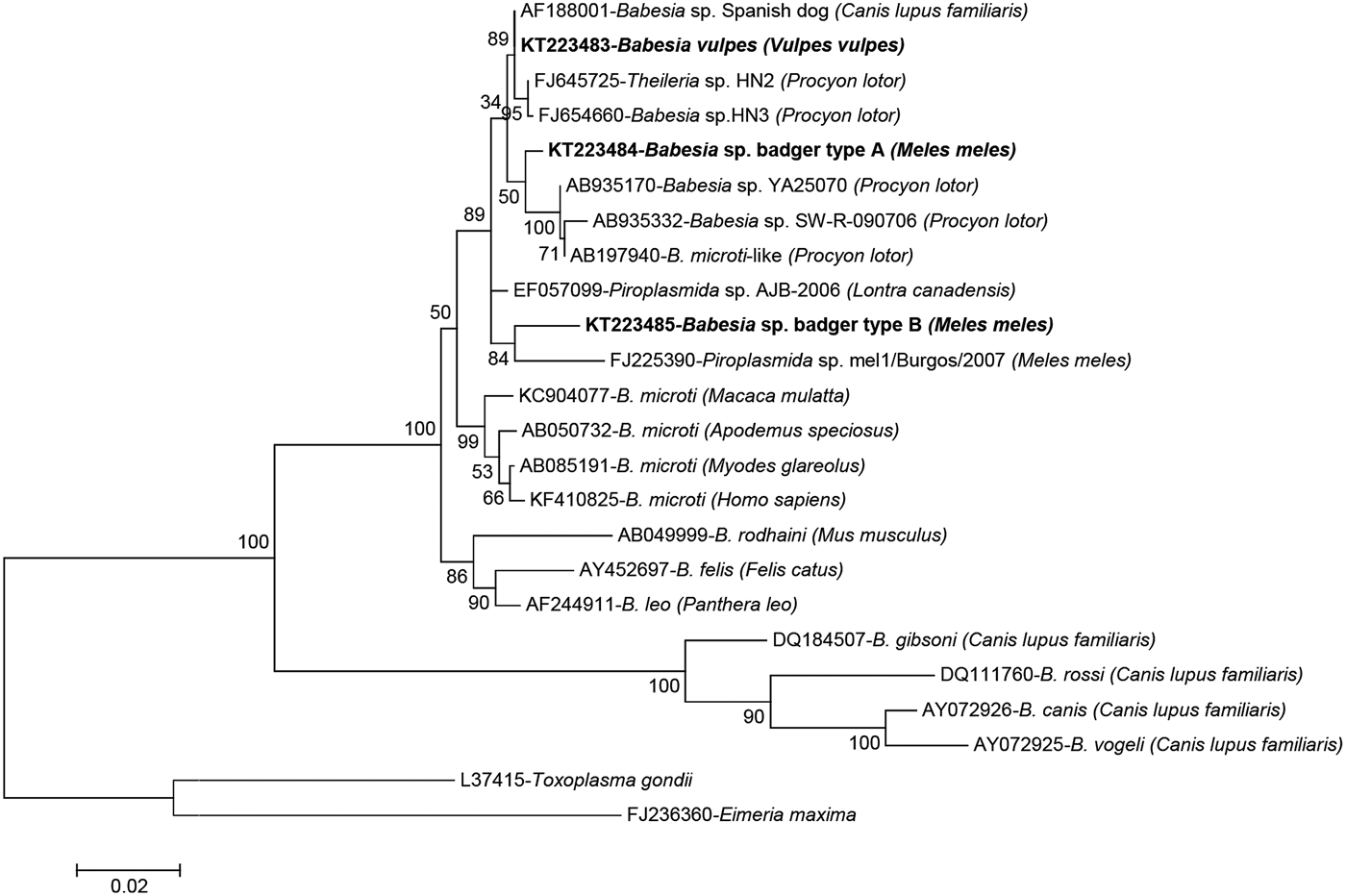
Fig. 1. Phylogenetic tree of the 18S rRNA gene inferred by maximum-likelihood analysis depicting the placement of Babesia spp. from wild carnivores derived from this study and representative neighbouring taxa. The tree with the highest log-likelihood is shown, which was inferred under the Tamura–Nei model with 5 gamma-distributed rate categories, allowing a percentage of sites to be held as invariable. All positions containing gaps and missing data were eliminated. There were a total of 1207 positions in the final dataset. Bootstrap values (1000 replicates) are indicated for each node. Each sequence is identified by its GenBank accession number, its definition as deposited in GenBank and the isolation host in brackets. The sequences described in this study are indicated in bold. Scale bar used was nucleotide substitutions per position.
Table 2. Sequencing analysis results

a One European badger harboured a co-infection with Cystoisospora sp. and Hepatozoon sp.
One hundred of the 238 animals externally examined for ticks harboured at least one tick (Table 1), with a maximum of 30 ticks collected in one badger. A total of 453 ticks of seven different species (44 were not identified to species level) were collected, 10.1% being larvae, 47.5% nymphs and 42.4% adults. Badgers (50.0%) and red foxes (45.8%) were the most parasitized species, the exception being American mink (Mustela vison) (100%, but only two specimens were examined). Ixodes hexagonus was the most abundant tick among Mustelidae and Canidae species, while I. ricinus was more abundant (37/38) in common genets (Genetta genetta). No statistically significant differences in piroplasm infection in relation with tick parasitization were found.
DISCUSSION
The present study provides evidence for piroplasm infection in wild carnivores from northern Spain, including red foxes, badgers and wild cats. Infection with B. vulpes, formerly known as T. annae (Baneth et al. Reference Baneth, Florin-Christensen, Cardoso and Schnittger2015), has been reported to cause severe illness in dogs (Camacho, Reference Camacho2006), and red foxes seem to be an important reservoir of this species in Spain and in other European and American countries (Gimenez et al. Reference Gimenez, Casado, Criado-Fornelio, de Miguel and Dominguez-Penafiel2009; Birkenheuer et al. Reference Birkenheuer, Horney, Bailey, Scott, Sherbert, Catto, Marr, Camacho and Ballman2010; Dezdek et al. Reference Dezdek, Vojta, Curkovic, Lipej, Mihaljevic, Cvetnic and Beck2010; Cardoso et al. Reference Cardoso, Cortes, Reis, Rodrigues, Simoes, Lopes, Vila-Vicosa, Talmi-Frank, Eyal, Solano-Gallego and Baneth2013; Najm et al. Reference Najm, Meyer-Kayser, Hoffmann, Herb, Fensterer, Pfister and Silaghi2014). In this investigation, B. vulpes was the only haemoparasite detected in red fox. It was identified in 17 of 48 analysed red foxes (35.4%), which represents a higher proportion than that reported in the neighbouring county of Burgos (1/5, 20.0%) (Gimenez et al. Reference Gimenez, Casado, Criado-Fornelio, de Miguel and Dominguez-Penafiel2009) but clearly lower than that found in Portugal in a larger study (63/91, 69.2%) (Cardoso et al. Reference Cardoso, Cortes, Reis, Rodrigues, Simoes, Lopes, Vila-Vicosa, Talmi-Frank, Eyal, Solano-Gallego and Baneth2013). Other Babesia species such as B. canis and B. gibsoni identified in red foxes (Penzhorn, Reference Penzhorn2006; Cardoso et al. Reference Cardoso, Cortes, Reis, Rodrigues, Simoes, Lopes, Vila-Vicosa, Talmi-Frank, Eyal, Solano-Gallego and Baneth2013) were not detected in any of the red foxes analysed in the study herein. Babesiosis by B. canis is frequently diagnosed in dogs in these two regions, but the first two clinical cases of dogs infected by B. vulpes have only recently been reported in Asturias (Miro et al. Reference Miro, Checa, Paparini, Ortega, Gonzalez-Fraga, Gofton, Bartolome, Montoya, Galvez, Mayo and Irwin2015), and as far as we know there are no published reports in the Basque Country. Conversely, B. vulpes infection is considered endemic in dogs in Northwestern Spanish regions such as Galicia (Camacho, Reference Camacho2006), whereas in Central and Southern Spain its prevalence is lower (Criado-Fornelio et al. Reference Criado-Fornelio, Martinez-Marcos, Buling-Sarana and Barba-Carretero2003). However, the observation of this parasite in blood smears is difficult, particularly when parasitaemia is low, and the disease might be underdiagnosed. Two new Babesia sp. badger sequence types were found in the European badger from both regions studied here, type A being five times more prevalent than type B. Both types were different to each other, to B. vulpes and to piroplasms described elsewhere, including the new piroplasm identified from badgers in Burgos (Gimenez et al. Reference Gimenez, Casado, Criado-Fornelio, de Miguel and Dominguez-Penafiel2009). In the phylogenetic analysis, the two Babesia sp. from badger clearly clustered with Babesia spp. that infect carnivores. Babesia sp. badger type B clearly branched with a babesia identified from a badger in Burgos (FJ225390) despite sharing only 96.5% homology in pairwise comparisons. The specific position of Babesia sp. badger type A in relation to the clade of B. vulpes and its sister clade of Babesia sp. from raccoon (Procyon lotor) could not be significantly established (low bootstrap values regardless of the method used to infer the tree; data not shown).
The four Cytauxzoon sp. sequences obtained from wild cats were identical or closely related to sequences from lynxes or domestic and wild cats from Europe (Criado-Fornelio et al. Reference Criado-Fornelio, Gónzalez-del-Río, Buling-Sarana and Barba-Carretero2004, Reference Criado-Fornelio, Buling, Pingret, Etievant, Boucraut-Baralon, Alongi, Agnone and Torina2009a ; Luaces et al. Reference Luaces, Aguirre, Garcia-Montijano, Velarde, Tesouro, Sanchez, Galka, Fernandez and Sainz2005; Gallusová et al. Reference Gallusová, Jirsová, Mihalca, Gherman, D'Amico, Qablan and Modrý2016). Cytauxzoon felis is a haemoparasite of felids that may cause severe clinical disease with high mortality (Meinkoth and Kocan, Reference Meinkoth and Kocan2005). Cytauxzoonosis is considered an emerging disease in the USA where it has been frequently reported in felids (Meinkoth and Kocan, Reference Meinkoth and Kocan2005). The bobcat (Lynx rufus) appears to be the natural reservoir of C. felis in North America. In Europe, presence of this haemoparasite was first described in 2005 in an Iberian lynx (Luaces et al. Reference Luaces, Aguirre, Garcia-Montijano, Velarde, Tesouro, Sanchez, Galka, Fernandez and Sainz2005). Until recently, only a few cases of Cytauxzoon infection had been reported, but in 2012 the first endemic focus in Europe was described in domestic cats in Italy, reporting a high prevalence of infection (23%) (Carli et al. Reference Carli, Trotta, Chinelli, Drigo, Sinigoi, Tosolini, Furlanello, Millotti, Caldin and Solano-Gallego2012). More recently, a high prevalence of Cytauxzoon infection was reported in wild cats (6/12) and in Eurasian lynx (4/4) in the Carpathian region of Romania (Gallusová et al. Reference Gallusová, Jirsová, Mihalca, Gherman, D'Amico, Qablan and Modrý2016). The similarly high proportion of positive wild cats found in our study (4/6) suggests that infection in wild felids in Europe might be more widespread than previously thought.
Besides these piroplasms, the detection of Hepatozoon sp. and Cystoisospora sp. in badgers (in one badger as a mixed infection) was an incidental finding that reflects only occurrence but not prevalence. However, to our knowledge this is the first description of Hepatozoon sp. in badger. Analysis of its 18S rRNA gene sequence showed that it was nearly identical (99.7%) to a new species, most closely related to, but distinct from Hepatozoon canis, described in pine martens in Scotland (Simpson et al. Reference Simpson, Panciera, Hargreaves, McGarry, Scholes, Bown and Birtles2005) and in Spain (Criado-Fornelio et al. Reference Criado-Fornelio, Bulling, Casado, Jimenez, Ruas, Wendt, Rosa-Farias, Pinheiro, Rey-Valeiron and Barba-Carretero2009b ), indicating that the same species could infect different free-ranging mustelids. Cystoisospora spp. are ubiquitous intestinal protists of wild and domestic carnivores that can also invade extra-intestinal organs such as spleen of definitive and paratenic host (Dubey, Reference Dubey2014). Badgers are commonly infected with Cystoisospora melis (Pellérdy, Reference Pellérdy1955), however, sequences of this coccidium are not available and published records are based on the morphology of oocysts isolated from badgers’ fecal samples (Anwar et al. Reference Anwar, Newman, Macdonald, Woolhouse and Kelly2000). In fact, only a few Cystoisospora spp. have been sequenced, C. melis not being among them. Ultimately, further studies are needed to estimate the true prevalence of these protists in wild carnivores.
Climatic and environmental conditions in Northern and Northwestern Spain allow for high burdens of some ixodid ticks (Barandika et al. Reference Barandika, Berriatua, Barral, Juste, Anda and García-Pérez2006). The most abundant tick species collected from both red foxes and badgers in this study was I. hexagonus, which is the potential vector of B. vulpes in the region (Camacho et al. Reference Camacho, Pallas, Gestal, Guitian, Olmeda, Telford and Spielman2003). The high proportion of I. hexagonus among ticks collected from badgers might indicate its possible role as vectors of the new babesias found in badgers in this study. The number of wild cats and collected ticks was too low to infer any conclusions concerning possible tick vectors of Cytauxzoon sp. On the other hand, and unlike the other tick-borne protists studied here, Hepatozoon transmission takes place by ingestion of the tick that acts as definitive host or by predation and ingestion of tissue forms from infected small wild mammals (Baneth, Reference Baneth2011).
In conclusion, the present study showed that B. vulpes is widespread among red foxes in northern Spain and demonstrated that badgers can harbour a high diversity of haemoparasite species, including the two different Babesia taxa described here for the first time, and different from B. vulpes and other babesia from carnivores. The presence of Hepatozoon sp. in Mustelidae is not novel (it has been reported in pine marten), but this is the first evidence of its presence in badgers specifically. The high proportion of wild cats infected with Cytauxzoon sp. found in the small sample analysed here requires further investigation. In summary, this study provided novel data on the different haemoparasites that can infect wild carnivores.
ACKNOWLEDGEMENTS
We thank Diputaciones Forales and hunters from the Basque Country and colleagues from SERIDA Animal Health Department and gamekeepers and other personnel from the Consejería de Fomento, Ordenación del Territorio y Medio Ambiente of Asturias, for providing animal samples. We are grateful to our colleagues Xeider Gerrikagoitia, Nekane Cortabarria and Ianire Astobiza for their laboratory assistance in DNA extraction at NEIKER. We would also like to thank Dr. Tomás Camacho (Laboratorio Lema & Bandín, Spain) for kindly providing us with control samples for B. vulpes (formerly T. annae). The authors would like to thank Dr N. Elguezabal (NEIKER) for revising the final version of the manuscript and improving the English style.
FINANCIAL SUPPORT
This work was supported by funding from INIA RTA2011-00008-C02-01/02 and the European Regional Development Fund (ERDF). This was not awarded to any specific author.
CONFLICT OF INTEREST
The authors declare no conflict of interest.




