Hair and eye color are two major features in determining an individual's appearance within a population. Both hair and eye color are highly heritable. Heritability estimates for hair color range from 61% to 100%, and for eye color similar high estimates are obtained (Brauer & Chopra, Reference Brauer and Chopra1980; Lin et al., Reference Lin, Mbarek, Willemsen, Dolan, Fedko and Abdellaoui2015; Zhu et al., Reference Zhu, Evans, Duffy, Montgomery, Medland and Gillespie2004). Linkage studies indicate quantitative trait loci (QTLs) on chromosomes 15q (Kayser et al., Reference Kayser, Liu, Janssens, Rivadeneira, Lao and van Duijn2008; Posthuma et al., Reference Posthuma, Visscher, Willemsen, Zhu, Martin and Slagboom2006), which contain the well-known pigment genes OCA2 and HERC2. Both hair and eye color are determined by gene variants present in the melanin pathway, including HERC2/OCA2, SLC24A4, and TYR (Candille et al., Reference Candille, Absher, Beleza, Bauchet, McEvoy and Garrison2012; Liu et al., Reference Liu, van Duijn, Vingerling, Hofman, Uitterlinden and Janssens2009; Sulem et al., Reference Sulem, Gudbjartsson, Stacey, Helgason, Rafnar and Magnusson2007; Zhang et al., Reference Zhang, Song, Liang, Nan, Zhang and Liu2013), though these genes may not explain all genetic variance in hair and eye color. Here, we focus on the question whether all genes affecting one of these visible traits also affect the other trait. To address this question, we estimated the genetic correlation between hair and eye color for common single nucleotide polymorphisms (SNPs) within a sample from the Dutch population using genomic restricted maximum likelihood (GREML) estimation for bivariate analyses (Lee et al., Reference Lee, Yang, Goddard, Visscher and Wray2012) as implemented in the Genome-wide Complex Trait Analysis (GCTA) software package (Yang et al., Reference Yang, Lee, Goddard and Visscher2011). The data came from unrelated individuals (N = 3,619) registered with the Netherlands Twin Register, which includes participants from all regions of the Netherlands. We explored the effect of including principal components (PCs) on the genetic correlation between hair and eye color. The first three PCs in the Dutch population significantly correlate with participants’ geographic location: PC1 with north–south, PC2 with east–west, and PC3 with center belt region of the Netherlands (Abdellaoui et al., Reference Abdellaoui, Hottenga, de Knijff, Nivard, Xiao and Scheet2013). Peripheral pigmentation traits such as eye, hair, and skin color show a close correspondence with latitudes in national to worldwide geographic regions: lower-pigmentation prevalence is found to be higher in high latitudes (Abdellaoui et al., Reference Abdellaoui, Hottenga, de Knijff, Nivard, Xiao and Scheet2013; Beleza et al., Reference Beleza, Santos, McEvoy, Alves, Martinho and Cameron2013). Bolk (Reference Bolk1908) reported in an article that early in the 20th century in the Netherlands, the northern part of the country was characterized by more blond hair and lighter eye color than the southern part of the country. As is evident from Table 1 using data from the Netherlands Twin Register collected around 2004, this is still the case roughly a 100 years later.
TABLE 1 The Distribution (in %) of Hair Color and Eye Color Across Dutch Provinces in the 1908 Sample Collected by Bolk (N=480,165) and for the 2004 Survey in Adult NTR Participants (N=7,661). We Report Three Classes as in the Original 1908 Paper, i.e., Blond Hair–Blue Eyes, Black Hair–Brown Eyes, and Red Hair

Note: aThis variable is Blond hair + blue eyes in the 1908 data and Blond hair + blue/gray eyes in the 2004 data.
bThe province of Flevoland consists of reclaimed land and did not yet exist in 1908.
Methods
Participants in the Netherlands Twin Register (van Beijsterveldt et al., Reference van Beijsterveldt, Groen-Blokhuis, Hottenga, Franic, Hudziak and Lamb2013; Willemsen et al., Reference Willemsen, Vink, Abdellaoui, den Braber, van Beek and Draisma2013) were included in this study based on the presence of self-reported data on natural hair and eye color and the presence of genotype data on an Illumina 370, 660, 1M or Affymetrix Perlegen-5.0, or 6.0 platform. There were 7,063 genotyped Dutch-ancestry participants, clustered in 3,407 families with data on eye color, and 6,965 genotyped individuals had data on both hair and eye color. For the genetic association analysis of eye color (see Supplementary material) all data were analyzed. For bivariate genetic analyses in GCTA, all unrelated individuals were selected, based on a genetic relatedness matrix (GRM) cut-off of 0.025 (Yang et al., Reference Yang, Lee, Goddard and Visscher2011). This left 3,619 individuals for the bivariate analyses, with a genetic relatedness equivalent to less than third or fourth cousin.
Age, sex, natural hair, and eye color were obtained from Adult NTR survey 7, which was collected in 2004 (Willemsen et al., Reference Willemsen, Vink, Abdellaoui, den Braber, van Beek and Draisma2013). Adult participants reported their own natural hair color from one of five options: ‘fair/blond’, ‘hazel’, ‘red/auburn’, ‘dark brown’, and ‘black’ and eye color with one of three options: ‘blue/gray’, ‘green/hazel’ and ‘brown’. The same questions on eye color and hair color were answered by adolescent (14- to 18-year-old) twins when they completed the Dutch Health and Behavior Questionnaire in 2005 or 2006 (van Beijsterveldt et al., Reference van Beijsterveldt, Groen-Blokhuis, Hottenga, Franic, Hudziak and Lamb2013). For the statistical analyses, we combined the black, light brown, and dark brown hair colors to ‘dark’, as only very few people reported a black hair color (Lin et al., Reference Lin, Mbarek, Willemsen, Dolan, Fedko and Abdellaoui2015). Written informed consent was obtained from all participants.
DNA extraction, purification, and genotype calling of the samples were performed at various points in time following the manufacturer's protocols and genotype calling programs (Lin et al., Reference Lin, Mbarek, Willemsen, Dolan, Fedko and Abdellaoui2015). For each platform, the individual SNPs were remapped on the build 37 (HG19), ALL 1000 Genomes Phase 1 imputation reference dataset (Auton et al., Reference Auton, Brooks, Durbin, Garrison, Kang and Korbel2015). SNPs that failed unique mapping and SNPs with an allele frequency difference over 0.20 with the reference data were removed. SNPs with a minor allele frequency (MAF) < 0.01 were also removed, as well as SNPs that were out of Hardy–Weinberg Equilibrium (HWE) with p < 10−5. The platform data were then merged into a single genotype set and the above SNP QC filters were reapplied. Samples were excluded from the data when their DNA was discordant with their expected sex or IBD status, the genotype missing rate was above 10%, the Plink F-inbreeding value was either larger than 0.10 or smaller than −0.10, or they were an ethnic outlier based on EIGENSTRAT PCs calculated from the 1000G imputed data (Auton et al., Reference Auton, Brooks, Durbin, Garrison, Kang and Korbel2015). Phasing of the samples and imputing cross-missing platform SNPs was done with MACH 1 (Li & Abecasis, Reference Li and Abecasis2006). The phased data were then imputed with MINIMAC to the 1000G reference. After imputation, SNPs were filtered, based on Mendelian error rate (>2%), a R 2 imputation quality value of <0.80, MAF <0.01 and a difference of more than 0.15 between the allele frequency and the reference (Howie et al., Reference Howie, Fuchsberger, Stephens, Marchini and Abecasis2012). We tested the effect of different platforms and removed SNPs showing platform effects. This was done by defining individuals on a specific platform as cases and the others as controls. If the allelic association between the specific platform allele frequency and the other platform's allele frequency was significant (p < 10−5) SNPs were removed. This left 5,987,253 SNPs, which were all used to construct a GRM.
A GRM based on autosomal SNPs was obtained from GCTA on the best-guess imputed data from Plink 1.07 (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira and Bender2007). The genetic correlation between the various dichotomous hair and eye color combinations (i.e., defined by whether a color was present or absent for the eyes and hair) was estimated using the GCTA bivariate analysis option (Lee et al., Reference Lee, Yang, Goddard, Visscher and Wray2012). Sex and age were used as covariates in the analyses. Next, we added three Dutch PCs calculated from the genetic data (Abdellaoui et al., Reference Abdellaoui, Hottenga, de Knijff, Nivard, Xiao and Scheet2013) as covariates, to explore the effect of ancestry-informative PCs on the analysis, since hair and eye color are likely related to the population diversity captured by these PCs.
Results
GWAS
The GWAS for hair color in this sample was published previously (Lin et al., Reference Lin, Mbarek, Willemsen, Dolan, Fedko and Abdellaoui2015) and the results from the GWAS for eye color are described in the Supplementary material. We replicated the known genetic variants for eye color including the HERC2 region for brown eye color (top SNP: rs74940492, OR = 0.09, p = 5.4E-8) and for blue/gray eye color (top SNP: rs2240202, OR = 13.55, p = 1.0E-47); TYR and SLC24A4 for blue/gray (top SNPs: rs4904871, OR = 0.71, p = 2.8E-13; rs67279079, OR = 0.70, p = 3.1E-11) and green/hazel eye color (top SNPs: rs4904871, OR = 1.52, p = 3.8E-20; rs67279079, OR = 1.49, p = 3.6E-10). Among these identified pigment genetic variants, we detected that HERC2 has pleiotropic effects on blond, brown, dark hair color and blue/gray, brown eye color, and SLC24A4 has pleiotropic effects on blond, brown, dark hair color and blue/gray and green/hazel eye color.
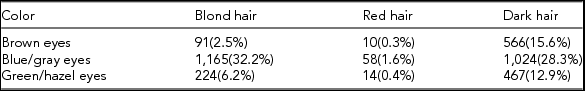
The phenotypic association of eye and hair colors confirms the two traits to be strongly related in our sample (χ 2-test with 4 degrees of freedom gave a p value < 2.2×10−16): people with blond and red hair are likely to have blue/gray eyes while people with dark hair are more likely to have brown eyes. The counts and frequencies of the hair and eye color phenotypes for the 3,619 individuals (1,401 males; age: 41.04±19.81 and 2,218 females; age: 39.13±17.17) are presented in Table 2(a), whereas Table 2(b) summarizes the information on the hair-eye color association from the much larger sample collected by Bolk (Reference Bolk1908), confirming the strong association in the Dutch population.
TABLE 2(a) Hair and Eye Color Counts and Percentages for Unrelated Genotyped Individuals of the Netherlands Twin Register

TABLE 2(b) Hair and Eye Color Counts and Percentages From Blom 1908

The genetic correlations between the hair and eye colors are presented in Table 3. Here, the same relation is shown as in the phenotypic description of the data, where the genes related to blond hair show a strong positive correlation with blue/gray colored eyes and a negative correlation with brown and green colored eyes. Due to the low prevalence of red hair color in our population, we do not detect any significant genetic overlap with any eye color. Finally, there is a clear and strong genetic overlap for brown eyes and a dark hair color.
TABLE 3 The Genetic Correlations Between Hair and Eye Color Estimated From Common SNPs (MAF > 0.01) Covering the Full Genome, Corrected for Age and Sex, Within Unrelated Individuals of the Netherlands Twin Register

Note: *One-sided p value < .05.
**One-sided p value < .01.
***One-sided p value < .001.
Likelihood ratio χ 2-test, df = 1, with the correlation fixed at 0.
When adding the first three genetic PCs that correlated with Dutch ancestry, the genetic correlations are reduced to zero (LRT = 0, p value = .5). This indicates that the genetic PCs of the Dutch population are capturing the overlapping genetic variance of eye and hair colors.
Discussion
Based on our analyses of genome-wide SNP data, there is a strong genetic overlap between eye and hair color within the Dutch population. This is in line with findings from previous molecular studies indicating that the same genes are involved in hair and eye color; for example, variants within the melanin producing pathway including HERC2, OCA2, SLC24A4, and TYR (Han et al., Reference Han, Kraft, Nan, Guo, Chen and Qureshi2008; Liu et al., Reference Liu, Wollstein, Hysi, Ankra-Badu, Spector and Park2010; Sulem et al., Reference Sulem, Gudbjartsson, Stacey, Helgason, Rafnar and Magnusson2007). We also conducted a GWAS for each of the two traits in the NTR population (see Lin et al., Reference Lin, Mbarek, Willemsen, Dolan, Fedko and Abdellaoui2015, for hair color and Supplementary material for eye color). The results confirmed the involvement of two genes, HERC2 and SLC24A4, in both hair color and eye color.
It is important to realize when studying eye and hair color that these phenotypes can be highly correlated with the genetic constitution of the population. Although the overall pigmentation prevalence has changed during the past 100 years (see Table 1: hyper-pigmentation traits are more prevalent in 2004), the distribution pattern of pigment traits following latitude is still the same. PCs representing Dutch ancestry and geographic location are likely to explain the largest part of the variability of human pigment traits. As shown here, the effect of population stratification and the true effects of genes on the two traits are closely linked, as PC1–PC3 also explained the genetic overlap between the traits. In our study, we only selected European Caucasian individuals based on the genetic PC projection and 1,000 genomes. Subsequently, three Dutch PCs were calculated in the remaining individuals to account for the population stratification of regions where people live in the Netherlands. However, these PCs also capture multiple traits that likely underwent simultaneous genetic divergence between (sub)populations, such as eye and hair color. When conducting gene finding studies or GCTA analyses, researchers should therefore be aware of the effects of ancestral population differences on the relationship between stratified traits.
Acknowledgments
BD Lin is supported by a Ph.D. grant (201206180099) from the China Scholarship Council. This study was supported by multiple grants from the Netherlands Organization for Scientific Research (NWO: 016-115-035, 463-06-001, 451-04-034), ZonMW (31160008, 911-09-032); Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); Biomolecular Resources Research Infrastructure (BBMRI–NL, 184.021.007), European Research Council (ERC-230374); Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL: 184.021.007). Genotyping was made possible by grants from NWO/SPI 56-464-14192, Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health, Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute, Sioux Falls (USA) and the National Institutes of Health (NIH R01 HD042157-01A1, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995).
Supplementary Material
To view supplementary material for this article, please visit https://doi.org/10.1017/thg.2016.85.