Introduction
Coxiella burnetii is an intracellular zoonotic pathogen responsible for Q fever in humans and coxiellosis in domestic and wild mammals. In humans, Q fever is associated with a wide clinical spectrum, from asymptomatic or mildly symptomatic seroconversion to fatal disease. Moreover, in humans acute and chronic Q fever are frequently misdiagnosed and underreported. Farm animals and pets are the main reservoirs of infection and the transmission to humans is mainly accomplished through inhalation of contaminated aerosols [Reference Angelakis and Raoult1]. In domestic ruminants, the infection may be asymptomatic or symptomatic. When symptomatic, it may cause epidemic abortion in sheep and goats as well as sporadic abortion, infertility and subclinical mastitis in cattle [Reference Barlow2–Reference De Biase4]. C. burnetii infection can cause significant economic losses in livestock and high health care costs related to diagnosis, long term therapies, hospitalization and working days lost [Reference Garner5, Reference Van Asseldonk, Prins and Bergevoet6]. When abortions occur, high concentrations of C. burnetii are found in placenta and birth products of infected animals. Still, the shedding can also take place during normal deliveries [Reference Rodolakis7]. The shedding dynamics differ among ruminants in terms of routes of excretion, load, duration and frequency. In cattle, the bacterium is shed almost exclusively in milk. In goats, it is shed mostly in milk, with a minority shedding it in vaginal mucus or faeces. Sheep result heavily infected and shed the bacterium in faeces, vaginal mucus and milk [Reference Rodolakis7–Reference Muleme9]; this could explain why human outbreaks of Q fever are more often related to ovine flocks than to bovine herds. In the 2007–2009 period, large community outbreaks of Q fever occurred in the Netherlands, with over 3500 notified cases in the Dutch population. Proximity to aborting small ruminants and the presence of a large number of susceptible humans were identified as the main causes of the Q fever outbreaks in humans [Reference Roest8]. Thus, the identification of shedders is crucial to avoid infections in humans and to prevent the diffusion among farmed animals. The identification of shedders through direct diagnostic tests is an expensive process that is not yet completely standardized. A fourfold rise in antibody titres could be used to accurately identify acutely infected animals as well, even if this approach is also costly and might not be practical with large sample sizes [Reference Muleme9]. Serological tests (indirect immunofluorescence assay – IFA, complement fixation test – CFT and enzyme-linked immunosorbent assay – ELISA) are recommended for seroprevalence studies. CFT has a weak sensitivity compared to other methods and no IFA is commercially available for ruminants to our knowledge, therefore ELISA tests are generally preferred also for practical reasons [Reference Sidi-Boumedine10, Reference Natale11]. Seropositivity to C. burnetii is not strongly correlated with the shedding of the bacterium. In fact, some shedders may be seronegative. Due to this, the serology cannot be used to estimate the real contamination rate of the herds, but it is a valuable tool for the screening of the herds and flocks [Reference Berri12–15]. The sensitivity and specificity of milk and blood ELISA are not significantly different, but blood ELISA is necessary for the studies involving non-lactating cattle [Reference Guatteo13, Reference Paul16]. Studies have been carried out on ruminants to define the relationship between the specific serological response phase (phase I/II) and the acute/chronic infection stage in analogy with humans. The attempts to classify the shedding pattern based on these studies allowed to classify some groups of animals but no satisfactory results have been achieved at the individual level so far [Reference Böttcher17–Reference Lucchese19]. Serosurveys were performed to evaluate the exposure to C. burnetii among healthy ruminants in Europe, US, Africa and Asia. These serosurveys reported prevalence at the animal-level ranging from 11% to 19.5% in sheep and 6.2% to 14.4% in cattle. The reported prevalence at the herd-level ranged from 38.7% to 74% in sheep, and from 16.7% to 71% in cattle [Reference Capuano, Landolfi and Monetti20–Reference Villari28]. In Italy, the data on Q fever prevalence in ruminants are mainly related to animals with reproductive disorders and particularly to those with abortion as the major clinical problem [Reference Masala29–Reference Natale32]. The Commission Implementing Regulation (EU) 2018/1882 included Q fever among the diseases enlisted within the ‘e’ category, which need surveillance and specific rules for notification and reporting as defined by the Regulation (EU) 2016/429 (‘Animal Health Law’). Nevertheless, the surveillance and reporting of C. burnetii in animals are not harmonized in EU. In Italy, the outbreak management is currently regulated by the Presidential Decree 320/1954. The Decree states that specific restrictive measures on farmed animals and milk products must be adopted only when human Q fever cases are related to the exposure to infected animals. In EU, Q fever is a mandatory notifiable disease in humans and all cases are reported through The European Surveillance System. The definition of ‘Q fever case’ is established precisely within the EU regulation, but the surveillance framework is not harmonized across Member States [33, 34]. In Italy, Q fever is a notifiable disease regulated under the Informative System for Infectious Diseases (DM 15/12/90). Q fever cases must be notified to the Local Public Health Authority and reported annually to the Regional Public Health Authority and to the Ministry of Health. Until 2015, Q fever cases were reported within the generic group of ‘rickettsial infections’. Since 2015, the overall surveillance system has been strengthened in the Lazio region to cope with the 2015–2016 Extraordinary Jubilee. Every single Q fever case was immediately reported to the Regional Health Authority by the Local Public Health authority. Few information on C. burnetii diffusion in domestic ruminants is available in the Lazio region, as the only existing data concern the differential diagnosis performed in the case of abortion. Furthermore, these data are likely underestimated due to the low notification rate of abortions by farmers. Recently, some polymerase chain reaction (PCR)-positive samples from aborted animals bred in the Lazio region were typed by Multispacer Sequence Typing (MST). The resulting MST genotypes (ST32 and ST12) were identical to those previously detected in clinical human samples from other countries [Reference Di Domenico35]. This evidence emphasizes the need for reliable information on C. burnetii diffusion in this area. Consolidated prevalence data are useful to quantify the exposure of the animal population to pathogens and represent the first step within the decision process of the health authorities. For this purpose, a cross-sectional study was carried out in extensively grazing cattle and sheep in the Lazio region, where this farming system is commonly practiced. The prevalence at animal- and herd-level of C. burnetii was estimated and the related risk factors were identified. Data on notified human Q fever cases in the area were also collected and described.
Methods
Study area and sampling
This serosurvey was carried out in the 2013–2014 period in the province of Rome (Metropolitan City). The province covers almost one-third of the territory of the Lazio region, central Italy, including the flat area of the Roman country, the Tiber Valley and several hilly and mountainous areas. With 5.4 km2 and 4.4 million inhabitants, the Metropolitan City includes the city of Rome, which is the most populated municipality in Italy and the fourth most populous city in the European Union. Rome is also the greenest city in Europe, with 63.8% of its territory covered by green areas. A portion of these areas is agricultural and pastureland, extending from centre to the borders of the Metropolitan City, where cattle and sheep are reared in an extensive grazing system. The beef cattle breeding scheme adopted in the area includes both purebred and mixed breed animals. Purebred animals represent the high-value breeding stock held under controlled conditions on permanent pastures; mixed breed animals which are bred for the commercial production of beef calves, are held under free grazing conditions on wider pastures. Cattle and sheep blood samples were collected by government veterinarians within the national eradication programme of brucellosis. Sera were separated by centrifugation for 10 min at 1000 × g and stored at −20 °C until examination at the Regional State Laboratory where the authors operate.
Serological tests
Cattle and sheep sera were analysed to detect anti-phase I and anti-phase II antibodies against C. burnetii by an ELISA prepared with C. burnetii strain isolated from ruminants (ID Screen Q Fever Indirect Multi-speciesIdvet, Grabels, France). In the internal validation sheet, the manufacturer declared a sensitivity of 100% (CI 95%: 88.65%–100%) and a specificity of 100% (CI 95%: 98.49%–100%). The results were expressed in an optical density sample/positive control (S/P) ratio, measured at 450 nm. The samples presenting an S/P percentage >50% were considered as positive; those with 40% ⩽ S/P% < 50% were considered as doubtful; otherwise samples were considered as negative. In this study, doubtful results were considered as negative.
Study population and study design
Data on the animal population were obtained from the National Animal Registry Database (Banca Dati Nazionale: BDN) defining a study population of 206 000 sheep belonging to 461 farms and 30 000 cattle from 1627 farms. Only extensive grazing animals were considered as target population for both species. The study population was considered as homogeneous for infection risk as they were all herds from extensive farming under similar managing conditions. A two-stage cluster sampling was performed, with herds as primary units and animals as secondary units, to estimate the prevalence of animals with a detectable level of antibodies against C. burnetii. The following assumptions were used for both species: 30% expected prevalence, 95% confidence level, ±5% or standard error and 0.2 intra-cluster correlation coefficient (low within-cluster homogeneity). The calculated minimum sample size for each species was at least 2194 animals from 73 clusters (herds). An a priori number of 30 animals was set to be tested in each herd. The herds and animals to be tested were selected from the list of samples stored during the 2013 and 2014 national brucellosis eradication programme, using a simple random sampling (SRS) method. As the herds were selected by SRS sampling a fixed number of animals which was not proportional to the herd size, the prevalence estimates (P adj) and 95% CI were adjusted using weights to account for the cluster sampling design employed using the following formula:
where P h stands for the proportion of positive individuals in each selected herd (No. positive/No. tested), M h stands for the number of animals present in each selected herd and ∑M h stands for the total number of animals present in all the tested herds [Reference Ferrari36].
The sampled animals were clinically healthy cattle and sheep, aged at least one year, with no history of previous vaccination against coxiellosis. Information on age, sex, breed, herd size and other species housed in the holdings were extrapolated from BDN for each randomly sampled cattle serum. The total number of farmed ruminants and the other animal species housed in the holdings were the only information available for sheep in BDN.
Statistical analysis
The prevalence at animal-level was separately calculated in cattle and sheep, considering the herd size as the weighting factor. The prevalence at herd-level was calculated for each species as the proportion of herds with at least one positive ELISA test on the total number of tested herds; 95% CI for prevalence were estimated by binomial distribution. The Chi square (χ 2) test was used to compare prevalence estimates between cattle and sheep. The following animal-level risk factors were considered: herd size (No. of cattle/sheep in the herd), species housed in the holding (cattle, sheep or both), age (in months), sex (male, female) and breed (mixed breed – MB, Maremmana – MRN, Charolaise – CHL, other breeds – other). The effect of the recorded risk factors on the outcome was assessed at animal- and herd-levels. Age and herd size were reported as medians and interquartile range (IQR, 75°–25° centile) after the Kolmogorov–Smirnov test for normality distribution. These two variables were divided into classes following the quantile classification method. To evaluate linearity, categories were created and the Log odds ratios (ORs) obtained from the univariate analysis performed were plotted against the midpoint of the category [Reference Dohoo, Martin and Stryhn37]. Discrete and qualitative data were reported as frequencies and percentages (%) respectively. The effect of the risk factors on the individual presence of detectable antibodies against C. burnetii was analysed using univariate logistic regression models. After excluding the associated variables (χ 2 test), the risk factors with a bivariate P value ⩽0.25 were included in a multivariate backward logistic model [Reference Hosmer and Lemeshow38]. The likelihood ratio χ 2 test was applied in order to select the best fitting model. The herd size and other animal species housed in the holdings were considered as risk factors for the herd-level analysis. The reference classes (ref) for risk factors were set as a herd size <30, age <37 months, male sex and MRN breed for cattle. The reference classes adopted for sheep were: herd size <176 and only-sheep housing. The strength of the association between the risk factors and the outcome was estimated using the ORs with 95% CI at animal- and herd-levels. A P value <0.05 was considered statistically significant. All statistical analyses were performed by Stata/SE version 12 for Windows (StataCorp LP, TX, USA).
Humans
Q fever cases in humans were notified by the doctors to the Local Public Health Authority. The Local Public Health Authority annually reported the notifications to the Regional Public Health Authority. Following the intensified notification flow after the 2015–16 Jubilee, every single case was reported also to Regional Public Health Authority. In accordance with the EU regulation, the cases were classified as: possible, probable or confirmed cases [33]. Demographic data, diagnostic criteria, date of symptom onset and risk factors were also collected and described for each case.
Mapping
ArcGIS 10.3® was used for mapping. The distribution of the holdings, weighted by the herd size, was used to build a kernel density map using the following parameters: 5 km radius (bandwidth), 1 km2 cell size and natural breaks classification (Jenks). The other layers used were: human cases, tested herds and not tested herds within 5 km buffers around the human cases.
Results
Cattle
A total of 2210 animals were tested for C. burnetii. The median age of the tested animals was 5.47 years (IQR 5.94), and 93.03% were female. The most frequent breeds were MB (55.7%) and MRN (15.57%). The prevalence at animal-level was 12.0% (95% CI 9.28–14.77) (Table 1). At the univariate analysis, the following animal-level risk factors were identified: age ≥67 months (OR 2.72, 95% CI 1.82–4.07), female sex (OR 2.28, 95% CI 1.11–4.71) and MB (OR 1.68, 95% CI 1.08–2.61). MRN breed was recognized as a protective factor. The presence of antibodies against C. burnetii in the animals did not result statistically associated neither with the herd size, nor with the presence of other farmed species in the holdings. In the multivariate analysis, only age and breed were maintained as significant risk factors within the best fitting model, since the effect of sex was lost in the first step of the backward deletion procedure. The animal-level risk factors confirmed in the best fitting model were: age classes 67–107 months (OR 2.79, 95% CI 1.86–4.18), >107 months (OR 2.07, 95% CI 1.36–3.14) and MB (OR 1.74, 95% CI 1.11–2.72) (Table 2).
Table 1. Animal-level and herd-level prevalence of C. burnetii in cattle and sheep

CI, confidence interval.
Table 2. Univariate and multivariate analyses of the animal-level prevalence of C. burnetii in cattle
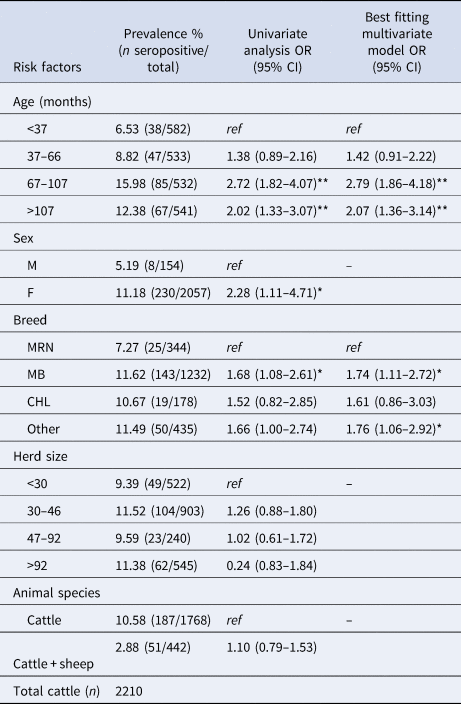
OR, odds ratio; CI, confidence interval; ref, reference category.
*P < 0.05; **P < 0.01.
A total of 92 herds were tested. The median herd size was 41.5 animals (IQR 57.75) and the median number of ruminants housed was equal to 49 (IQR 102.5). Seventy-three herds (79.4%) housed only cattle and 19 (20.6%) both cattle and sheep. Overall, 6511 heads were housed within the selected herds, accounting for 21.7% of the target population living in the study area. The prevalence at herd-level was 68.5% (95% CI 57.9–77.7) (Table 1). An increasing risk for herds to result positive for antibodies against C. burnetii was observed in relation to the herd size. Notably at the univariate analysis, herd size >92 showed about seven times higher risk (OR 6.88, 95% CI 1.67–28.37) compared to the reference class. No association was found between the presence of at least one animal with detectable antibodies against C. burnetii and the presence of other farmed animal species in the same holding (Table 3).
Table 3. Univariate analysis of of the herd-level prevalence of prevalence of C. burnetii in cattle

OR, odds ratio; CI, confidence interval; ref, reference category.
*P < 0.05; **P < 0.01.
Sheep
A total of 2873 animals were tested for C. burnetii. The prevalence at animal-level was 37.8% (95% CI 33.3–42.3) (Table 1). At the univariate analysis, the risk of being seropositive was double in sheep belonging to flocks >600 animals (odds ratio (OR) 2.04, 95% CI 1.63–2.56) compared to the reference class (flock size <176). No statistical association was observed between the presence of detectable antibodies against C. burnetii in the animals and the presence of other farmed animal species in the holdings.
A total of 94 flocks were tested. The median flock size was 360 animals (IQR 392.5) and the median number of ruminants was 365.5 (IQR 446.75). Sixty-four flocks (68.1%) housed only sheep and 30 (31.9%) housed both sheep and cattle. Overall, 42 596 sheep were farmed within the tested flocks, accounting for 20.7% of the target population living in the study area. The prevalence at herd-level was 87.2% (95% CI 78.8–93.2) (Table 1). The univariate analysis showed that the probability that a flock was positive for antibodies against C. burnetii did not depend neither from the flock size, nor from the presence of other animal species housed in the holding.
Prevalence in cattle and sheep
The weighted prevalence at animal-level was three times higher in sheep compared to cattle (37.8% vs. 12.0%; χ 2 = 270.10, P < 0.001); the prevalence at herd-level resulted significantly higher in sheep than in cattle (87.2% vs. 68.5%; χ 2 = 9.52, P < 0.01).
Humans
Seven confirmed human Q fever cases were reported. Six cases were localized in the rural area surrounding the city of Rome and one in the province of Viterbo, close to the border of the province of Rome (Fig. 1). The date of onset of clinical disease was from November 2016 to November 2017. All cases had positive serology following clinical manifestations. Five cases were male and one female. The proximity to small ruminant holdings was explicitly referred to as risk factor in three cases. None of the cases was reported to work in the sampled farms. In five out of seven cases there was at least one exposed herd within a 5 km buffer. One case was living 6.8 km far from exposed herds. The case falling within the province of Viterbo was living 0.327 km far from the nearest herd. All cases fell within cells having a density of ruminants from 9.8 to 156.9 heads per km2. The distance to the nearest herd is comprised from 0.169 to 1.024 km and in two cases the nearest herd was an exposed herd (Table 4). The infection source was not identified.

Fig. 1. Human Q fever cases and tested herds in the study area; not tested herds within 5 km buffers around the human cases.
Table 4. Human Q fever cases in the study area

E, exposed.
a The nearest herd is an exposed herd. b6.8 km far.
Discussion
In this study, the prevalence at animal-level of C. burnetii was 12.0% in healthy cattle. Other studies carried out in Italy reported prevalence ranging from 14.4% to 22% in healthy cows, whereas a 44.9% prevalence was reported in cows with abortion [Reference Capuano, Landolfi and Monetti20, Reference Cabassi30]. We found that the risk of testing positive for antibodies against C. burnetii increased in mixed breed cattle compared to the reference class (MRN breed). This result is consistent with the different exposure to this pathogen that apparently occurred, by considering the beef cattle breeding scheme adopted in the study area. MRN breed animals are purebred subjects, belonging to the high-value breeding stock used for the production of commercial female breeders. The purebred animals are kept under more controlled conditions consisting of fenced permanent grazing pastures, ensuring levels of higher biosecurity. Conversely, the mixed breed animals belonging to the category of breeders, used for the commercial production of beef calves, are held in conditions of free grazing on wider pastures where contacts with animals from different herds often occur. For this reason they could be probably more at risk for the transmission of the infection. In a previous study, the Friesian breed resulted a risk factor for the presence of antibodies against C. burnetii [Reference Mccaughey23]. In the present study, the probability of having detectable antibodies against C. burnetii increased with the age (>67 months). This result is in line with other studies indicating that the odds of being infected are higher in older animals [Reference Mccaughey23, Reference Alvarez39]. This finding could be explained with a higher probability of being exposed to the pathogen over the years. In a study carried out in Italy, the prevalence at animal-level of C. burnetii was found to be influenced by herd management. In detail, it was 19.6% in those herds housed in winter and unhoused in spring; 13.2% in permanently housed animals and 1.9% in unhoused cattle [Reference Capuano, Landolfi and Monetti20]. In Spain, Ruiz-Fons et al. reported a 6.7% prevalence at animal-level in a semi-extensive grazing system [Reference Ruiz-Fons22]. By contrast, in this study the prevalence at animal-level was higher than those observed by the above-mentioned authors. The higher prevalence of C. burnetii observed in the area could be due to different breeding methods, different densities of receptive animal species, and to the eco-environmentally favourable conditions for the exposure to the pathogen. Due to the lack of information currently available for these potential risk factors, further ad hoc investigations should be performed to compare the prevalence of C. burnetii among different herd management conditions in the study area. In this study, the prevalence at herd-level was 68.5% in cattle. In Italy, 4.4% herd-level prevalence was estimated using CFT [Reference Martini, Baldelli and Paulucci De Calboli40]. In Spain 43.0% herd-level prevalence was reported in cattle [Reference Ruiz-Fons22]. The wide range of the seroprevalence reported by those studies could be due to the distinct epidemiological scenarios. Nevertheless, the prevalence reported was also affected by the performances of the assays employed. With regards to the ELISA employed in this study, the manufacturer reported a 100% sensitivity (CI 95%: 88.65%–100%) and a 100% specificity (CI 95%: 98.49%–100%). In the worst-case scenario, considering the lower confidence limit of the sensitivity (88.65%) provided by the manufacturer, the prevalence estimates provided in our study were probably underestimated compared to the true prevalence. By converse, the lower confidence limit of the specificity (98.49%) suggested a low probability of occurrence of false negative results, in accordance with what is well known for a highly specific test such as the CFT. Thus, an overestimation of prevalence was unlikely to have occurred. Anyway, by considering the lower confidence limits of both specificity and sensitivity of the ELISA test employed, the positive predictive value obtained was expected to be very high, considering also the high seroprevalence observed. CFT is not so suitable for serological studies since it fails to detect cases when anti-complementary substances are present in sera. Moreover, in ruminants some antibodies are not revealed by CFT because only IgG1 antibodies are known to fix the complement. Furthermore, CFT titers may be reduced due to the presence of IgG2 and IgM antibodies which can suppress complement fixation by IgG1 antibodies [Reference Sidi-Boumedine10, Reference Rousset41]. In our study, the risk of having at least one animal with detectable antibodies against C. burnetii was seven times higher in herds with more than 92 heads. This result is consistent with the hypothesis that animals from larger herds were more likely to come into contact with infected individuals. In particular, an increasing animal density could result in a higher probability of being directly exposed to C. burnetii during the deliveries. Antibodies against C. burnetii were detected in sheep from an extensive farming system with 37.8% prevalence at animal-level. Recent surveys carried out on healthy sheep in northwest Italy and in Sicily reported 16.3% and 18.0% individual prevalence respectively [Reference Rizzo27, Reference Villari28]. Nine percent prevalence at animal-level was estimated using a Nine Mile ELISA test in sheep with abortion [Reference Masala29]. In this study, the risk of being exposed to C. burnetii resulted double for sheep belonging to flocks with >600 animals. The higher risk of being seropositive for animals from larger flocks may be related to the greater number of lambing females which increases the total population at risk and, subsequently, the risk of introduction and transmission of this pathogen. In our study, the prevalence at herd-level was 87.2% in sheep. The probability of having at least one positive animal did not depend neither from the flock size, nor from the presence of other animal species housed in the holding. This finding is not in agreement with those studies reporting the flock size as herd-level risk factor for C. burnetii [Reference Rizzo27]. The prevalence at animal-level of C. burnetii resulted three times higher in sheep compared to cattle (37.8% vs. 12.0%); the prevalence at herd-level was higher in sheep than in cattle (87.2% vs. 68.5%) as well. These findings could be explained with the higher infection load in sheep and with the several ways of shedding that occurs in this species, resulting in a substantial exposure to the pathogen [Reference Rodolakis7, Reference Rousset14, Reference Guatteo42]. Furthermore, the gregarious aptitude of sheep enhances the contacts among animals and so the probability of being exposed to C. burnetii through direct and indirect routes also in an extensive grazing system. Our results highlight the considerable diffusion of C. burnetii in the study area. It was unexpected to find such a high prevalence in an extensive grazing system. This evidence led us also to hypothesize a possible involvement of ticks in the epidemiology of the pathogen in the area. Notoriously, ticks represent a secondary route of infection of C. burnetii in ruminants. The vector competence has been demonstrated for many hard and soft tick species even if the vector capacity could be low in field conditions. Conversely, the tick-borne transmission of the bacterium seems to be more efficient in wild animals. In addition, the recent discovery of Coxiella-like bacteria, endosymbionts of several tick species, imply the possibility that they could be transmitted to vertebrates during the blood meal and misidentified as C. burnetii when vertebrates are screened using either direct or indirect tests [Reference Duron43, Reference Angelakis44]. Moreover, pastures and vegetation can be frequently shared by domestic and wild animals in an extensive grazing system, thus facilitating the transmission of a huge variety of pathogens from the wild reservoirs, through the direct or indirect route [Reference Cantas45–Reference Barlozzari47]. Sheep were confirmed to be the most exposed species at both animal- and herd-levels. However, the seroprevalence observed in cattle suggests also a potential involvement of this species in the circulation of the bacterium in the study area. The presence of domestic ruminants substantially exposed to an air-borne zoonotic pathogen indicates that also humans can be potentially exposed and become infected. In the study area, five out of seven confirmed human Q fever cases had at least one exposed herd within a 5 km buffer. The distance to the nearest herd is comprised from 0.169 to 1.024 km and in two cases the nearest herd is an exposed herd. In a recent review, the highest risk of infection for humans occurred within 5 km of infected farms in rural areas [Reference Clark and Soares Magalhães48]. Even if the infection source was not identified in any of those cases, the possibility of C. burnetii circulating in the livestock and human population in the study area cannot be overlooked. These findings suggest the need to implement surveillance and control systems based on a ‘One Health’ approach, involving the systematic notification of cases in both humans and animals. The integration between veterinary and human surveillance will be crucial to understand the spread of this zoonosis and to support the adoption of appropriate control measures. Collaborative surveillance platforms where cases and outbreaks are reported could facilitate the epidemiological investigations and define the infection risk. Efforts were made in the Lazio region to develop such a platform following the collaboration between the IZSLT and the Seresmi. However, further studies are needed in the study area to collect and characterize the strains circulating in ruminants and to compare them with those isolated from humans, through a Multiple-Locus Variable number tandem repeat Analysis/MST approach or Next-Generation Sequencing. Such an approach would be useful to a better understanding of the epidemiology of C. burnetii and to trace the strains in the case of outbreaks.
Acknowledgements
The authors wish to thank Giuseppina Lionetti for her assistance and skillful management of sample storage. Thanks are due to Patrizia Gradito for English editing.
Financial support
This study was supported by the Italian Ministry of Health (grant code IZS LT 13/10 RC).
Conflict of interest
None.






