Impact statement
Precision oncology, one of the most promising applications of precision medicine, uses molecular and genetic information to customise cancer treatments, considering the individual characteristics of each patient’s tumour. To further advance the field, precision oncology increasingly incorporates knowledge of cancer heterogeneity, on both spatial and temporal levels. Addressing these complexities with modern precision radiotherapy and systemic therapies is the key to targeting all cancer cell subpopulations. The future vision of precision oncology involves continuous advancements in technological and analytical methods, leading to further treatment personalisation. This progress will ultimately contribute to a paradigm shift in cancer care to improve patient outcomes significantly. Access to advanced tools should be improved in terms of availability and affordability while addressing the need for routine genomic profiling across various regions of primary and metastatic tumours to understand cancer heterogeneity comprehensively.
Introduction
Precision medicine is a novel approach to treatment and prevention that tailors strategies to the unique characteristics of individual patients, including their genetics, environment and lifestyle. It differs from conventional evidence-based medicine, which generally relies on average clinical benefits in the studied populations (Tonelli and Shirts, Reference Tonelli and Shirts2017; Blackstone, Reference Blackstone2019). Precision medicine is supported by advances in technology and medical research, such as using genomic sequencing and big data analysis to identify individualised treatment options.
The decision-making process in precision medicine is based on predictive biomarkers, which offer insights into the underlying molecular mechanisms of tumorigenesis and allow the identification of potential therapeutic targets. In clinical settings, biomarkers can predict which patients are most likely to benefit from specific therapies, optimise treatment efficacy and reduce toxicity. Further, biomarkers enable early cancer detection and treatment monitoring, thereby increasing its efficacy. In essence, biomarkers are transformative tools of personalised medicine, driving more accurate, effective and safer cancer treatments (Slikker, Reference Slikker2018).
The two most commonly used markers are prognostic and predictive biomarkers. A prognostic biomarker is a clinical or biological indicator that offers insights into the probable health outcome of an individual patient, such as disease recurrence or death, regardless of the treatment pursued. In turn, a predictive biomarker signifies the potential advantage to the patient, resulting from a specific treatment (Sechidis et al., Reference Sechidis, Papangelou, Metcalfe, Svensson, Weatherall and Brown2018). Other biomarkers include predisposing biomarkers, indicating the potential for developing a disease (Califf, Reference Califf2018) and pharmacogenomic biomarkers, informing about the drug efficacy and toxicity based on the underlying genetic composition (Lauschke et al., Reference Lauschke, Milani and Ingelman-Sundberg2017). The United States Food and Drug Administration and the National Institutes of Health published the Biomarkers, EndpointS and other Tools (BEST) resource, describing the extensive list of biomarkers used in translational science (Cagney et al., Reference Cagney, Sul, Huang, Ligon, Wen and Alexander2018).
Precision oncology is a concept that customises oncological care based on unique patient genomics and clinical, genetic, proteomic, transcriptomic or phenotypic tumour features (de and Ashworth, Reference de Bono and Ashworth2010; Collins and Varmus, Reference Collins and Varmus2015). Precision oncology has achieved unprecedented advancements through rigorous scientific evidence and extensive computational analyses (Mirnezami et al., Reference Mirnezami, Nicholson and Darzi2012). However, challenges such as accurate data interpretation, precise patient stratification and the development of successful targeted therapies for specific genomic aberrations require further efforts (Prasad et al., Reference Prasad, Fojo and Brada2016).
To overcome these obstacles, precision oncology requires innovative clinical trial designs that account for patient and tumour heterogeneity and the dynamic nature of cancer evolution (Chen and Snyder, Reference Chen and Snyder2013). Integrating precision oncology into clinical practice is a key goal of the Precision Medicine Initiative, which was launched by the US government in 2015.
In the present article, we discuss the impact of tumour and patient heterogeneity on treatment outcomes in solid tumours oncology and explore how precision systemic therapies and radiotherapy can mitigate these obstacles. The analysis will focus on scrutinising pivotal studies, such as the Molecularly Aided Stratification for Tumour Eradication Research (MASTER) (Horak et al., Reference Horak, Klink, Heining, Gröschel, Hutter, Fröhlich, Uhrig, Hübschmann, Schlesner, Eils, Richter, Pfütze, Geörg, Meißburger, Wolf, Schulz, Penzel, Herpel, Kirchner, Lier, Endris, Singer, Schirmacher, Weichert, Stenzinger, Schlenk, Schröck, Brors, von Kalle, Glimm and Fröhling2017), the National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH) trial (Flaherty et al., Reference Flaherty Keith, Gray, Chen, Li, Patton, Hamilton, Williams, Mitchell, Iafrate, Sklar, Harris, McShane, Rubinstein, Sims, Routbort, Coffey, Fu, Zwiebel, Little, Marinucci, Catalano, Magnan, Kibbe, Weil, Tricoli, Alexander, Kumar, Schwartz, Meric-Bernstam, Lih, McCaskill-Stevens, Caimi, Takebe, Datta, Arteaga, Abrams, Comis, O’Dwyer and Conley2020 and other pertinent research, to better understand customised cancer therapy. We also present current investigative endeavours and interdisciplinary collaborations to optimise cancer therapy in all patients, regardless of their genetic makeup (Topol, Reference Topol2014; Jameson and Longo, Reference Jameson and Longo2015; Figure 1). The examples provided here should be considered illustrative, as no comprehensive literature analysis on this topic was attempted.

Figure 1. Timeline showing the highlights of clinical precision medicine. ALK, anaplastic lymphoma kinase; ALL, acute lymphoblastic leukaemia; BCa, breast carcinoma; BRAF, v-Raf murine sarcoma viral oncogene homologue B’ BRCA, BReast CAncer gene; CAR-T, chimeric antigen receptor T-cell therapy; CML, chronic myeloid leukaemia; CR, complete response; ER+, oestrogen receptor-positive; HER2, human epidermal growth factor receptor-2; KRAS, Kirsten rat sarcoma virus; MET, hepatocyte growth factor receptor; MMRd, mismatch repair deficiency; MSI-H, high microsatellite instability; NGS, next generation sequencing; NSCLC, non-small cell lung cancer; PD-1, programmed death receptor-1; RET, Ret Proto-Oncogene; VEGF, vascular endothelial growth factor.
Systemic therapies
Systemic therapies, which involve drugs circulating throughout the body, are fundamental to cancer treatment (Chabner and Roberts, Reference Chabner and Roberts2005). Precision oncology has revolutionised systemic therapies by better allocating standard chemotherapy and has paved the way for specific targeted therapies (Schwaederle et al., Reference Schwaederle, Zhao, Lee, Eggermont, Schilsky, Mendelsohn, Lazar and Kurzrock2015). However, cancer heterogeneity, both spatial and temporal, highly impacts the effectiveness of these therapies (Greaves and Maley, Reference Greaves and Maley2012). As a result, one of the major challenges in oncology is customising systemic therapies for each patient and tumour characteristics (Leichsenring et al., Reference Leichsenring, Horak, Kreutzfeldt, Heining, Christopoulos, Volckmar, Neumann, Kirchner, Ploeger, Budczies, Heilig, Hutter, Fröhlich, Uhrig, Kazdal, Allgäuer, Harms, Rempel, Lehmann, Thomas, Pfarr, Azoitei, Bonzheim, Marienfeld, Möller, Werner, Fend, Boerries, von Bubnoff, Lassmann, Longerich, Bitzer, Seufferlein, Malek, Weichert, Schirmacher, Penzel, Endris, Brors, Klauschen, Glimm, Fröhling and Stenzinger2019).
Tumour heterogeneity
Whereas cytotoxic chemotherapy is essential for many malignancies, it is generally recognised as a one-size-fits-all approach, which may not be optimal for patients with genetically diverse tumours. Precision oncology considers tumour genetic heterogeneity, thus can improve the efficacy of standard treatments, identify druggable targets for specific tumours and select patients who are more likely to benefit from customised treatments (Massard et al., Reference Massard, Michiels, Ferté, Le Deley, Lacroix, Hollebecque, Verlingue, Ileana, Rosellini, Ammari, Ngo-Camus, Bahleda, Gazzah, Varga, Postel-Vinay, Loriot, Even, Breuskin, Auger, Job, De Baere, Deschamps, Vielh, Scoazec, Lazar, Richon, Ribrag, Deutsch, Angevin, Vassal, Eggermont, André and Soria2017).
The relationship between specific genomic alterations, genetic inter- and intratumour heterogeneity and the effectiveness of cancer treatment has been well established (Schwaederle et al., Reference Schwaederle, Zhao, Lee, Eggermont, Schilsky, Mendelsohn, Lazar and Kurzrock2015; McGranahan and Swanton, Reference McGranahan and Swanton2017). Tumours with certain genetic alterations differ in their susceptibility to classical cytotoxic chemotherapy. For example, mutations in the TP53, KRAS, PTEN or RB1 genes are associated with resistance to chemotherapy (Custodio et al., Reference Custodio, González-Larriba, Bobokova, Calles, Alvarez, Cuadrado, Manzano and Díaz-Rubio2009; Perrone et al., Reference Perrone, Bossi, Cortelazzi, Locati, Quattrone, Pierotti, Pilotti and Licitra2010), BRCA1 and BRCA2 mutations denote chemosensitivity to platinum compounds (Pennington et al., Reference Pennington, Walsh, Harrell, Lee, Pennil, Rendi, Thornton, Norquist, Casadei, Nord, Agnew, Pritchard, Scroggins, Garcia, M-C and Swisher2014), and MGMT methylation in glioblastoma is associated with a better response to temozolomide (Stupp et al., Reference Stupp, Mason, van den Bent, Weller, Fisher, Taphoorn, Belanger, Brandes, Marosi, Bogdahn, Curschmann, Janzer, Ludwin, Gorlia, Allgeier, Lacombe, Cairncross, Eisenhauer and Mirimanoff2005).
Knowledge of genetic tumour heterogeneity has been extensively used in targeted cancer therapies (Figure 2). If druggable, genetic alterations are primarily used as therapeutic targets; however, many also serve as predictive markers for treatment effectiveness. In colorectal cancer, cetuximab, which is a chimeric antibody against the epidermal growth factor receptor (EGFR), is effective only against wild-type rat sarcoma (RAS) family oncogenes (Van Cutsem et al., Reference Van Cutsem, Köhne, Hitre, Zaluski, Chang Chien, Makhson, D’Haens, Pintér, Lim, Bodoky, Roh, Folprecht, Ruff, Stroh, Tejpar, Schlichting, Nippgen and Rougier2009; Douillard et al., Reference Douillard, Oliner, Siena, Tabernero, Burkes, Barugel, Humblet, Bodoky, Cunningham, Jassem, Rivera, Kocákova, Ruff, Błasińska-Morawiec, Šmakal, Canon, Rother, Williams, Rong, Wiezorek, Sidhu and Patterson2013). Conversely, in lung cancer, EGFR tyrosine kinase inhibitors are less effective in patients with coexisting TP53 (Aggarwal et al., Reference Aggarwal, Davis, Mick, Thompson, Ahmed, Jeffries, Bagley, Gabriel, Evans, Bauml, Ciunci, Alley, Morrissette, Cohen, Carpenter and Langer2018; Sun et al., Reference Sun, Ren, Chen, Lan, Yan, Yang, Wang, Wang, Yanwei, Li, Zhang, Yanyang, Wang, Pan and Jiang2023) or KRAS mutations (Massarelli et al., Reference Massarelli, Varella-Garcia, Tang, Xavier, Ozburn, Liu, Bekele, Herbst and Wistuba2007), which can activate alternative signalling pathways bypassing the EGFR pathway. In breast cancer, human epidermal growth factor receptor-2 (HER2) inhibitors are widely used to treat patients with HER2-overexpressing or HER2-amplified tumours, but they are less effective in patients with coexisting mutations in fibroblast growth factor receptor-1 (FGFR1) or receptor-2 (FGFR2) genes (Hanker et al., Reference Hanker, Garrett, Estrada, Moore, Ericsson, Koch, Langley, Singh, Kim, Frampton, Sanford, Owens, Becker, Groseclose, Castellino, Joensuu, Huober, Brase, Majjaj, Brohée, Venet, Brown, Baselga, Piccart, Sotiriou and Arteaga2017). FGFR alterations correlate with resistance to several targeted and standard therapies across different malignancies (Babina and Turner, Reference Babina and Turner2017), while the mechanistic and prognostic role of FGFR1–4 protein overexpression remains equivocal (Piasecka et al., Reference Piasecka, Braun, Kitowska, Mieczkowski, Kordek, Sadej and Romanska2019).
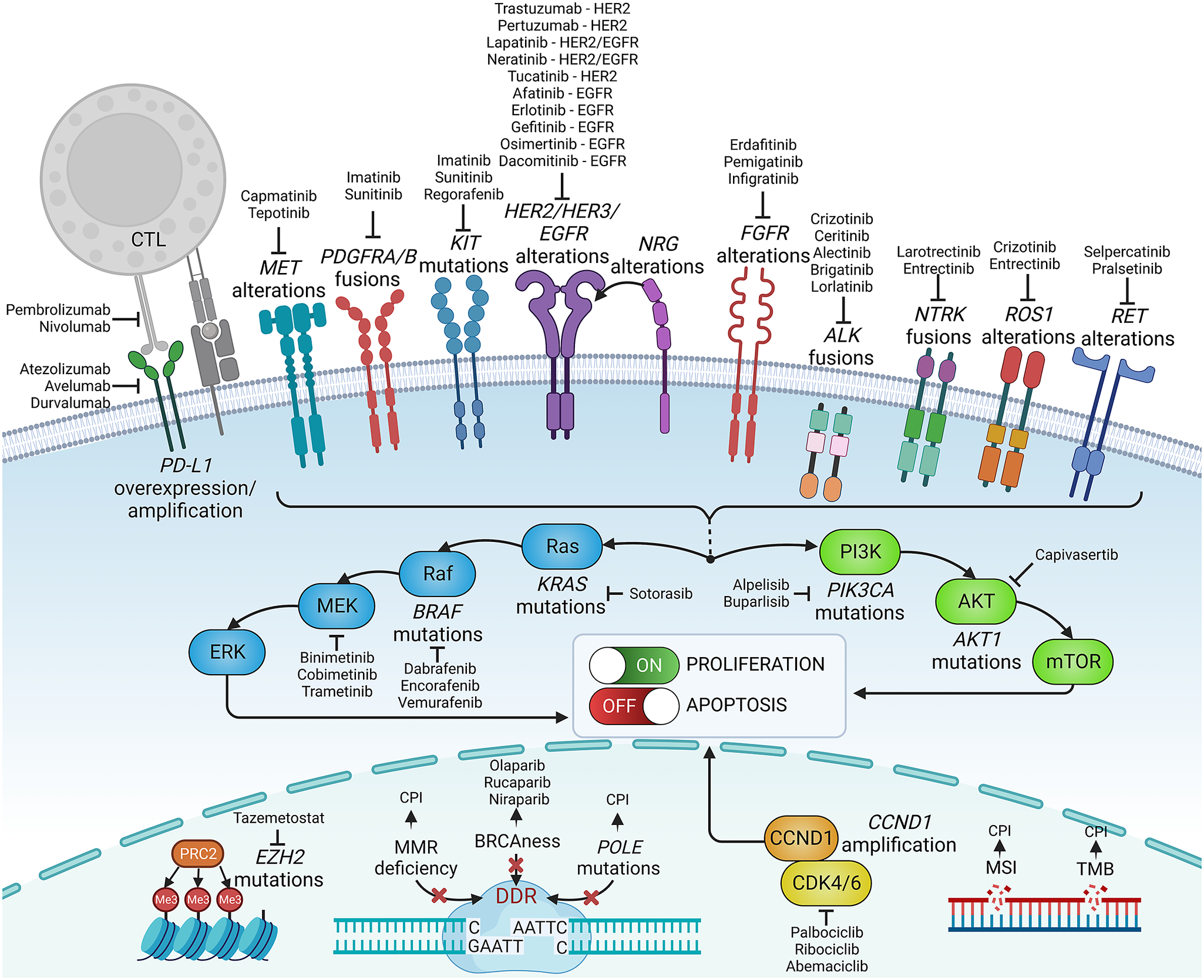
Figure 2. Examples of precision medicine biomarkers used in oncologic practice, together with respective targeted therapies approved in patients harbouring such lesions. CPI, checkpoint inhibitor; CTL, cytotoxic lymphocyte; DDR, DNA damage response; MMR, mismatch repair; MSI, microsatellite instability; PD-L1, programmed death-ligand 1; TMB, tumour mutational burden.
Some genetic alterations are druggable only in specific tumours, whereas others can be targeted across biologically and clinically different malignancies. The inhibitors of cyclin-dependent kinases 4 and 6 (CDK4/6) are effective and routinely administered to treat advanced hormone receptor-positive, HER2-negative breast cancer, though CDK4/6 alterations are not a hallmark of these cancers and do not predict the effectiveness of this therapy (Suski et al., Reference Suski, Braun, Strmiska and Sicinski2021; Cristofanilli et al., Reference Cristofanilli, Rugo, Im, Slamon, Harbeck, Bondarenko, Masuda, Colleoni, DeMichele, Loi, Iwata, O’Leary, André, Loibl, Bananis, Liu, Huang, Kim, Lechuga Frean and Turner2022). Furthermore, CDK4/6 inhibitors are inefficient in liposarcomas harbouring the amplification of CDK4/6 and murine double minute 2 (MDM2) genes (Sbaraglia et al., Reference Sbaraglia, Bellan and Dei Tos2021). Other examples are ivosidenib, an isocitrate dehydrogenase-1 (IDH1) inhibitor, and enasidenib, an IDH2 inhibitor, which effectively targets relapsed or refractory acute myeloid leukaemia with IDH1/2 mutations (Cerchione et al., Reference Cerchione, Romano, Daver, DiNardo, Jabbour, Konopleva, Ravandi-Kashani, Kadia, Martelli, Isidori, Martinelli and Kantarjian2021) but that are not effective in gliomas bearing these mutations. In turn, some targeted therapies, for example, entrectinib, which targets neurotrophic tyrosine receptor kinase (NTRK) fusions, and ROS oncogene 1 (ROS1) rearrangements, are effective across different solid tumour types, including lung cancer, colorectal cancer and thyroid cancer (Doebele et al., Reference Doebele, Drilon, Paz-Ares, Siena, Shaw, Farago, Blakely, Seto, Cho, Tosi, Besse, Chawla, Bazhenova, Krauss, Chae, Barve, Garrido-Laguna, Liu, Conkling, John, Fakih, Sigal, Loong, Buchschacher, Garrido, Nieva, Steuer, Overbeck, Bowles, Fox, Riehl, Chow-Maneval, Simmons, Cui, Johnson, Eng, Wilson and Demetri2020; Drilon et al., Reference Drilon, Siena, Dziadziuszko, Barlesi, Krebs, Shaw, de Braud, Rolfo, Ahn, Wolf, Seto, Cho, Patel, Chiu, John, Goto, Karapetis, Arkenau, Kim, Ohe, Li, Chae, Chung, Otterson, Murakami, Lin, Tan, Prenen, Riehl, Chow-Maneval, Simmons, Cui, Johnson, Eng, Wilson and Doebele2020). Similarly, V-raf murine sarcoma viral oncogene homologue B1 (BRAF) and mitogen-activated protein kinase (MEK) inhibitors were recently approved with a tumour-agnostic indication for unresectable or metastatic solid tumours harbouring the BRAF V600E mutation (Gouda and Subbiah, Reference Gouda and Subbiah2023).
Immune-oriented therapies, such as immune checkpoint inhibitors (ICIs) or chimeric antigen receptor T-cells (CAR-T), have revolutionised cancer treatment. However, correctly identifying good responders remains challenging. Genetic heterogeneity in tumours may elicit variable responses to ICIs. Malignancies with a high tumour mutational burden (TMB) and neoantigen load are more responsive to ICIs. Patients with high-TMB non-small cell lung cancer (NSCLC) or melanoma achieve significant improvements in survival with ICIs compared with those with low TMB (Ning et al., Reference Ning, Liu, Wang, Li, Xu and Wei2022; Ricciuti et al., Reference Ricciuti, Wang, Alessi, Rizvi, Mahadevan, Li, Polio, Lindsay, Umeton, Sinha, Vokes, Recondo, Lamberti, Lawrence, Vaz, Leonardi, Plodkowski, Gupta, Cherniack, Tolstorukov, Sharma, Felt, Gainor, Ravi, Getz, Schalper, Henick, Forde, Anagnostou, Jänne, Van Allen, Nishino, Sholl, Christiani, Lin, Rodig, Hellmann and Awad2022). However, intratumour or intersite (primary vs. metastatic foci) heterogeneity leading to spatial neoantigen expression variability might result in the escape of certain subclones from immune surveillance (McGranahan and Swanton, Reference McGranahan and Swanton2017). Different tumour types (e.g., colorectal or endometrial cancers) with microsatellite instability or mismatch repair deficiency are highly responsive to ICIs (Cercek et al., Reference Cercek, Lumish, Sinopoli, Weiss, Shia, Lamendola-Essel, El Dika, Segal, Shcherba, Sugarman, Stadler, Yaeger, Smith, Rousseau, Argiles, Patel, Desai, Saltz, Widmar, Iyer, Zhang, Gianino, Crane, Romesser, Pappou, Paty, Garcia-Aguilar, Gonen, Gollub, Weiser, Schalper and Diaz2022; O’Malley et al., Reference O’Malley, Bariani, Cassier, Marabelle, Hansen, De Jesus Acosta, Miller, Safra, Italiano, Mileshkin, Xu, Jin, Norwood and Maio2022), whereas tumours with some mutations may be ICI resistant. For instance, EGFR-mutant NSCLCs are less sensitive to ICIs than wild-type EGFR (Mazieres et al., Reference Mazieres, Drilon, Lusque, Mhanna, Cortot, Mezquita, Thai, Mascaux, Couraud, Veillon, Van den Heuvel, Neal, Peled, Früh, Ng, Gounant, Popat, Diebold, Sabari, Zhu, Rothschild, Bironzo, Martinez-Marti, Curioni-Fontecedro, Rosell, Lattuca-Truc, Wiesweg, Besse, Solomon, Barlesi, Schouten, Wakelee, Camidge, Zalcman, Novello, Ou, Milia and Gautschi2019). Melanomas with overactive WNT/β-catenin signalling are less infiltrated by T-cells and, thus, less susceptible to ICIs (Spranger et al., Reference Spranger, Bao and Gajewski2015).
Cancers with high levels of intrinsic heterogeneity, which can be defined as the presence of different genetic clones within a single tumour, are usually less likely to benefit from chemotherapy and targeted therapies (McGranahan and Swanton, Reference McGranahan and Swanton2017) because of the presence of drug-resistant subclones within the tumour that can contribute to rapid relapse after the initial response to therapy (Burrell et al., Reference Burrell, McGranahan, Bartek and Swanton2013; Almendro et al., Reference Almendro, Cheng, Randles, Itzkovitz, Marusyk, Ametller, Gonzalez-Farre, Muñoz, Russnes, Helland, Rye, Borresen-Dale, Maruyama, van Oudenaarden, Dowsett, Jones, Reis-Filho, Gascon, Gönen, Michor and Polyak2014). Computational modelling and in situ analyses have shown that genetic and phenotypic heterogeneity can greatly affect tumour evolution during chemotherapy and treatment outcomes (Almendro et al., Reference Almendro, Cheng, Randles, Itzkovitz, Marusyk, Ametller, Gonzalez-Farre, Muñoz, Russnes, Helland, Rye, Borresen-Dale, Maruyama, van Oudenaarden, Dowsett, Jones, Reis-Filho, Gascon, Gönen, Michor and Polyak2014). Recent advances in genomics and single-cell sequencing have shed light on the molecular mechanisms underlying tumour heterogeneity, paving the way for the development of novel personalised therapeutic strategies (Dagogo-Jack and Shaw, Reference Dagogo-Jack and Shaw2018; Ramón et al., Reference Ramón, Cajal, Sesé, Capdevila, Aasen, De Mattos-Arruda, Diaz-Cano, Hernández-Losa and Castellví2020; Labrie et al., Reference Labrie, Brugge, Mills and Zervantonakis2022). Further development of precision medicine for systemic anticancer therapies heavily relies on better understanding and addressing intratumour heterogeneity. However, because of diagnostic limitations, intratumour heterogeneity cannot yet be routinely exploited in guiding treatment options. It is expected that circulating tumour DNA (ctDNA) and single-cell analysis techniques may enable a detailed characterisation of tumour cell populations and better inform personalised treatment strategies (Nath and Bild, Reference Nath and Bild2021; Tivey et al., Reference Tivey, Church, Rothwell, Dive and Cook2022). ctDNA dynamic profiling allows for real-time monitoring of tumour evolution and adapting treatment strategies as the tumour mutates and evolves. Recently, ctDNA-guided therapy was shown to be beneficial in patients with NSCLC and colorectal cancer (Jee et al., Reference Jee, Lebow, Yeh, Das, Namakydoust, Paik, Chaft, Jayakumaran, Rose Brannon, Benayed, Zehir, Donoghue, Schultz, Chakravarty, Kundra, Madupuri, Murciano-Goroff, Tu, Xu, Martinez, Wilhelm, Galle, Daly, Yu, Offin, Hellmann, Lito, Arbour, Zauderer, Kris, Ng, Eng, Preeshagul, Victoria Lai, Fiore, Iqbal, Molena, Rocco, Park, Lim, Li, Tong-Li, De Silva, Chan, Diakos, Itchins, Clarke, Pavlakis, Lee, Rekhtman, Chang, Travis, Riely, Solit, Gonen, Rusch, Rimner, Gomez, Drilon, Scher, Shah, Berger, Arcila, Ladanyi, Levine, Shen, Razavi, Reis-Filho, Jones, Rudin, Isbell and Li2022; Tie et al., Reference Tie, Cohen, Lahouel, Lo, Wang, Kosmider, Wong, Shapiro, Lee, Harris, Khattak, Burge, Harris, Lynam, Nott, Day, Hayes, McLachlan, Lee, Ptak, Silliman, Dobbyn, Popoli, Hruban, Lennon, Papadopoulos, Kinzler, Vogelstein, Tomasetti and Gibbs2022).
Precision medicine approaches may considerably improve cancer treatment outcomes, provided that the complex interplay between tumour genetics and response to systemic treatment is better understood. Basic and translational studies are essential for identifying next-generation predictive biomarkers. Novel clinical trial designs, such as basket-type trials assessing the druggability of specific targets across different tumour types, and umbrella-type trials evaluating the efficacy of specific or various targeted therapies in specific cancer diagnoses (Figure 3 and Table 1), may prompt the development of new tailored therapies (Subbiah, Reference Subbiah2023).
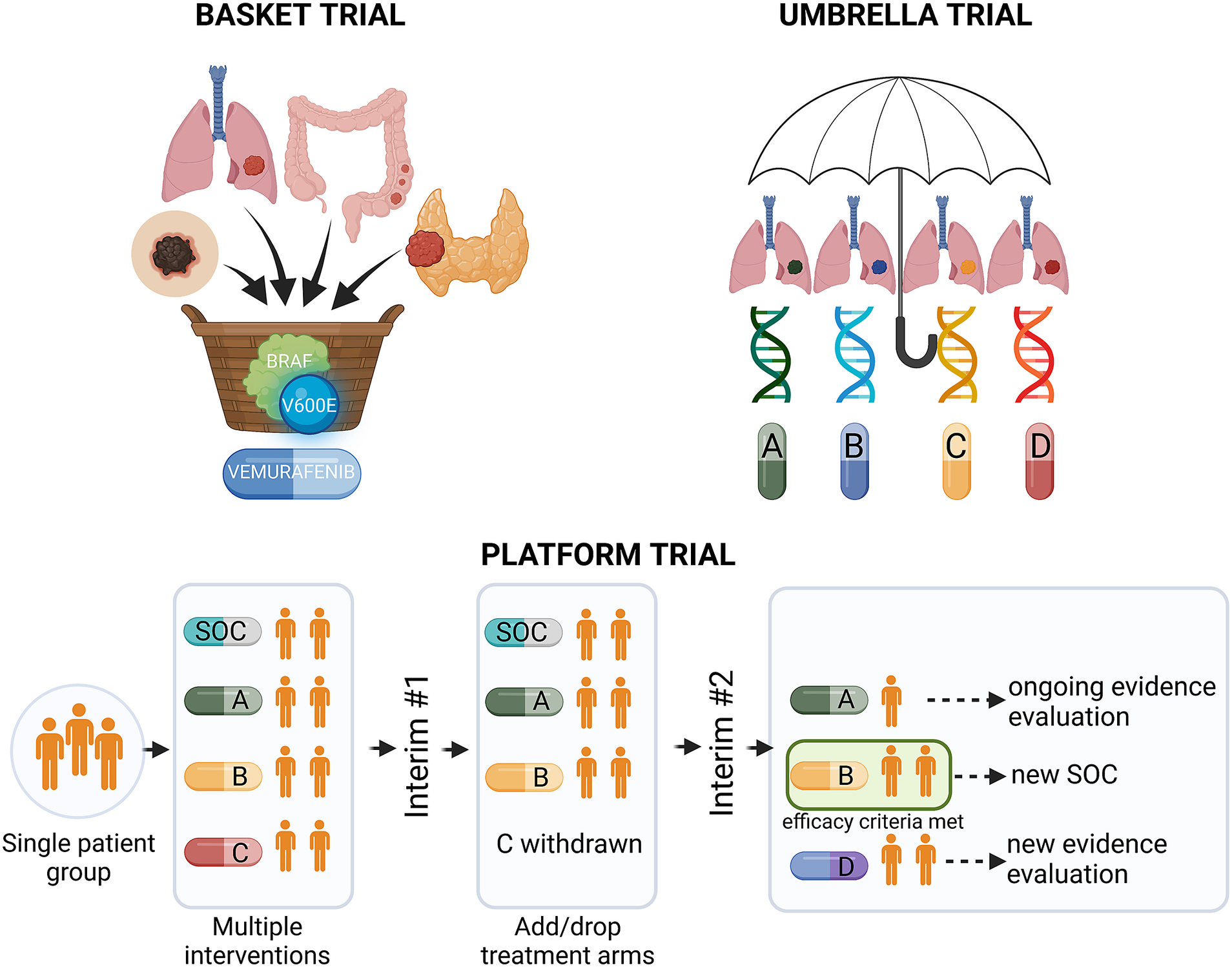
Figure 3. Types of precision medicine clinical trials. BRAF, v-Raf murine sarcoma viral oncogene homologue B; SOC, standard of care.
To compile the data presented in Tables 1 and 2 (please see below), we employed a multi-pronged search strategy. Firstly, a targeted search was conducted on PubMed using the following query: ("precision medicine"[Title/Abstract] OR “targeted therapy”[Title/Abstract] OR “personalised medicine”[Title/Abstract]) AND “clinical trial”[Publication Type] AND ("2013/01/01"[PDAT]: “2023/12/31”[PDAT]). This query was designed to yield articles classified as “clinical trials” focusing on “precision medicine,” “targeted therapy,” or “personalised medicine,” and published between January 1, 2013, and December 31, 2023. In addition to PubMed, we supplemented the articles with data from the clinical trials registry ClinicalTrials.gov and information gathered from sessions at the European Society for Medical Oncology (ESMO) Meetings and American Society of Clinical Oncology (ASCO) Annual Meetings held between 2018 and 2023. These conferences are recognised platforms that regularly feature key developments in precision medicine trials in oncology.
Table 1. Selected clinical trials investigating personalised cancer therapies

ALK, anaplastic lymphoma kinase; CBR, clinical benefit rate; CI, confidence interval; ctDNA, circulating tumour DNA; DCR, disease control rate; DMFS, distant metastasis-free survival; DNA, deoxyribonucleic acid; EGFR, epidermal growth factor receptor; FDA, Food and Drug Administration; FFPE, formalin-fixed, paraffin-embedded; ICD – O, International Classification of Diseases for Oncology; IHC, immunohistochemistry; KRAS, Kirsten Rat Sarcoma Viral oncogene homologue; MSI, microsatellite instability; MTB, molecular tumour board; NGS, next-generation sequencing; NR, not reported; NSCLC, non-small cell lung cancer; OR, odds ratio; ORR, objective response rate; OS, overall survival; PD-L1, programmed death-ligand 1; PFS, progression-free survival; PFSr, PFS interval associated with molecularly informed therapy (PFS2) divided by the PFS interval associated with the last prior systemic therapy (PFS1); RNA, ribonucleic acid; RNA-seq, RNA sequencing; TMB, tumour mutational burden; VEGF, vascular endothelial growth factor; VEGFR, vascular endothelial growth factor receptor; WES, whole exome sequencing; WGS, whole genome sequencing.
Table 2. Summary of selected clinical studies investigating radiosensitivity-predicting genomic signatures
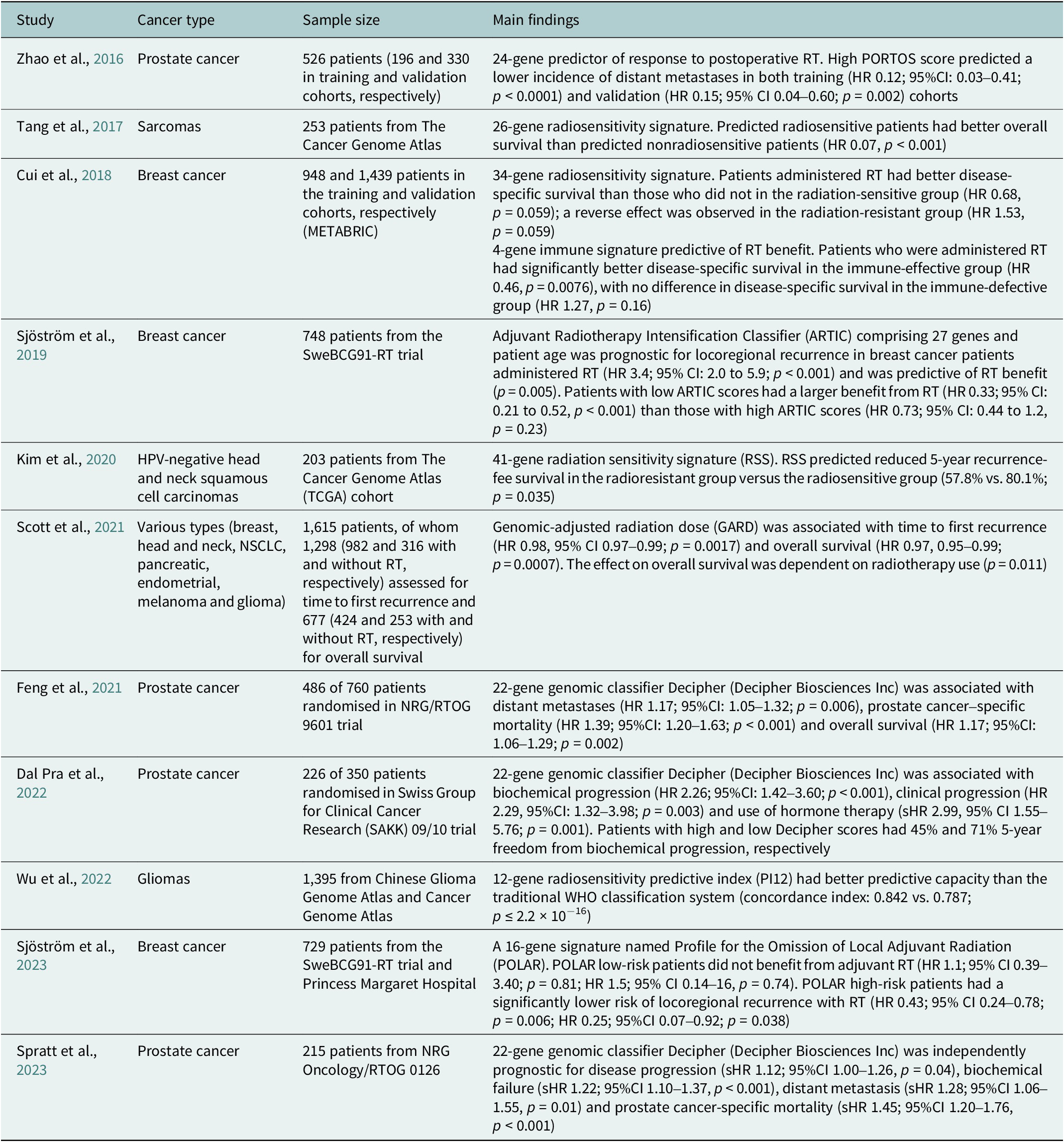
95%CI, 95% confidence interval; HR, hazard ratio; sHR, subdistribution hazard ratio.
Germline heterogeneity
Response and adverse reactions to systemic therapies can vary substantially between individuals. This variability has been attributed mainly to the inherited genomic variants that inactivate protein-coding genes (Karczewski et al., Reference Karczewski, Francioli, Tiao, Cummings, Alföldi, Wang, Collins, Laricchia, Ganna, Birnbaum, Gauthier, Brand, Solomonson, Watts, Rhodes, Singer-Berk, England, Seaby, Kosmicki, Walters, Tashman, Farjoun, Banks, Poterba, Wang, Seed, Whiffin, Chong, Samocha, Pierce-Hoffman, Zappala, O’Donnell-Luria, Minikel, Weisburd, Lek, Ware, Vittal, Armean, Bergelson, Cibulskis, Connolly, Covarrubias, Donnelly, Ferriera, Gabriel, Gentry, Gupta, Jeandet, Kaplan, Llanwarne, Munshi, Novod, Petrillo, Roazen, Ruano-Rubio, Saltzman, Schleicher, Soto, Tibbetts, Tolonen, Wade, Talkowski, Neale, Daly and MacArthur2020). The therapy may be impacted on several levels, including direct drug–target interactions, drug metabolism (including drug activation and removal) and downstream effects (e.g., DNA damage). Hence, considering patient pharmacogenomics can improve treatment outcomes, decrease toxicity and reduce costs.
The drug-metabolising enzyme known as cytochrome P450 2D6 (CYP2D6) is the most thoroughly examined and variable polymorphic enzyme (Zhou and Lauschke, Reference Zhou and Lauschke2022). Its deficiency is inherited through an autosomal recessive trait, and individuals carrying this alteration are classified as poor metabolisers. However, the remaining subjects (extensive metabolisers) display considerable variability in their enzymatic activity (Bertilsson et al., Reference Bertilsson, Dahl, Dalén and Al-Shurbaji2002). The gene encoding the CYP2D6 protein is highly polymorphic, with over 100 allelic variants described to date (Gaedigk et al., Reference Gaedigk, Ingelman-Sundberg, Miller, Leeder, Whirl-Carrillo and Klein2018). Specific genetic variations in the CYP2D6 gene can cause altered activity of the cytochrome P450 2D6 enzyme, which is involved in the metabolism of tamoxifen (a selective oestrogen modulator used to treat breast cancer). Individuals with reduced CYP2D6 activity (e.g., *4, *5 and *6 alleles) account for up to 10% of patients (Ingelman-Sundberg, Reference Ingelman-Sundberg2004; Crews et al., Reference Crews, Gaedigk, Dunnenberger, Leeder, Klein, Caudle, Haidar, Shen, Callaghan, Sadhasivam, Prows, Kharasch and Skaar2014). These individuals have a lower efficacy of tamoxifen than those with normal activity (Lim et al., Reference Lim, Ju Lee, Seok Lee, Sook Lee, I-J and Ro2007; Schroth et al., Reference Schroth, Antoniadou, Fritz, Schwab, Muerdter, Zanger, Simon, Eichelbaum and Brauch2007; Goetz et al., Reference Goetz, Suman, Hoskin, Gnant, Filipits, Safgren, Kuffel, Jakesz, Rudas, Greil, Dietze, Lang, Offner, Reynolds, Weinshilboum, Ames and Ingle2013). Likewise, the ultrarapid metabolisers (e.g., with *1xN or *2xN alleles, about 1–2% of patients) show lower endoxifen (a tamoxifen metabolite) concentrations and worse outcomes compared with patients with normal CYP2D6 activity (Wegman et al., Reference Wegman, Vainikka, Stål, Nordenskjöld, Skoog, L-E and Wingren2005; Schroth et al., Reference Schroth, Antoniadou, Fritz, Schwab, Muerdter, Zanger, Simon, Eichelbaum and Brauch2007; Crews et al., Reference Crews, Gaedigk, Dunnenberger, Leeder, Klein, Caudle, Haidar, Shen, Callaghan, Sadhasivam, Prows, Kharasch and Skaar2014).
Similarly, variations in the DPYD gene (e.g., *2A or *13), which encodes the dihydropyrimidine dehydrogenase, an enzyme involved in the metabolism of fluoropyrimidines, such as 5-fluorouracil and capecitabine, can lead to reduced enzyme activity and an increased risk of severe toxicity (Amstutz et al., Reference Amstutz, Farese, Aebi and Largiadèr2009; Offer et al., Reference Offer, Fossum, Wegner, Stuflesser, Butterfield and Diasio2014; Henricks et al., Reference Henricks, Lunenburg, de Man, Meulendijks, Frederix, Kienhuis, Creemers, Baars, Dezentjé, Imholz, Jeurissen, Portielje, Jansen, Hamberg, Ten Tije, Droogendijk, Koopman, Nieboer, van de Poel, Mandigers, Rosing, Beijnen, Werkhoven, van Kuilenburg, van Schaik, Mathijssen, Swen, Gelderblom, Cats, Guchelaar and JHM2018).
Finally, the UGT1A1 gene, which encodes for the uridine diphosphate glucuronosyltransferase 1A1 enzyme, is involved in the metabolism of irinotecan (a topoisomerase I inhibitor); variations reducing its activity (including *6, *27 and *28) may cause severe toxicity, including neutropenia, diarrhoea or infection (Iyer et al., Reference Iyer, Das, Janisch, Wen, Ramírez, Karrison, Fleming, Vokes, Schilsky and Ratain2002; Innocenti et al., Reference Innocenti, Undevia, Iyer, Chen, Das, Kocherginsky, Karrison, Janisch, Ramírez, Rudin, Vokes and Ratain2004; Marcuello et al., Reference Marcuello, Altés, Menoyo, Del Rio, Gómez-Pardo and Baiget2004; Xu et al., Reference Xu, Tang, Qu, Keyoumu, Zhou and Tang2016).
Genomic polymorphisms in drug targets may also affect interactions with drugs. Several HER2 gene variants have been shown to impact the effectiveness of trastuzumab (a monoclonal antibody targeting HER2) in HER2-positive breast cancer patients. The F117L variant, which is located in the extracellular domain of the HER2 protein, impairs trastuzumab binding by approximately threefold compared with wild-type HER2. Decreased binding affinity is attributed to the introduction of a leucine residue, which causes a steric hindrance and disrupts the protein conformation at the binding site (Gaborit et al., Reference Gaborit, Larbouret, Vallaghe, Peyrusson, Bascoul-Mollevi, Crapez, Azria, Chardès, Poul, Mathis, Bazin and Pèlegrin2011). In turn, a variant within the HER2 intracellular kinase domain (T798I) leads to increased kinase activity, conferring resistance to lapatinib (Bose et al., Reference Bose, Kavuri, Searleman, Shen, Shen, Koboldt, Monsey, Goel, Aronson, Li, Ma, Ding, Mardis and Ellis2013). Another example of EGFR polymorphism is R521K (rs2227983), which may decrease the response to cetuximab in patients with metastatic colorectal cancer (Graziano et al., Reference Graziano, Ruzzo, Loupakis, Canestrari, Santini, Catalano, Bisonni, Torresi, Floriani, Schiavon, Andreoni, Maltese, Rulli, Humar, Falcone, Giustini, Tonini, Fontana, Masi and Magnani2008). As a result, patients who carry this allele experience a lower incidence of skin toxicity during cetuximab treatment (Klinghammer et al., Reference Klinghammer, Knödler, Schmittel, Budach, Keilholz and Tinhofer2010; Fernández-Mateos et al., Reference Fernández-Mateos, Seijas-Tamayo, Mesía, Taberna, Pastor Borgoñón, Pérez-Ruiz, Adansa Klain, Vázquez Fernández, Del Barco Morillo, Lozano, González Sarmiento and Cruz-Hernández2016).
Finally, variants affecting the efficiency of DNA damage repair may affect the response and toxicity of numerous drugs, including platinum agents, alkylating agents, topoisomerase II inhibitors, antimetabolites and poly-ADP ribose polymerase inhibitors. The rs3212986 variant (C8092A) of the ERCC1 gene, which is a component of the nucleotide excision repair pathway, is associated with a poor response to platinum agents in NSCLC and ovarian cancer (Zhou et al., Reference Zhou, Gurubhagavatula, Liu, Park, Neuberg, Wain, Lynch, Su and Christiani2004; Krivak et al., Reference Krivak, Darcy, Tian, Armstrong, Baysal, Gallion, Ambrosone and DeLoia2008). Similarly, the rs13181 (L751G) single nucleotide variant (SNV) in the XPD gene is associated with the poor efficacy of platinum agents in NSCLC (Park et al., Reference Park, Stoehlmacher, Zhang, Tsao-Wei, Groshen and Lenz2001).
Radiotherapy
Radiotherapy is a vital component of cancer treatment, applicable in around 60% of patients (Citrin, Reference Citrin2017). Until recently, radiotherapy was prescribed on the empirical basis of a one-fits-all approach, assuming a similar response to the same radiation dose. Recent advances in precision medicine have enabled the use of more targeted and personalised radiotherapy regimens tailored to the specific characteristics of individual patients and their tumours. Cancer heterogeneity poses a significant challenge for radiotherapy, as it can cause variable tumour responses and the emergence of radioresistant cell populations. Precision radiotherapy, by considering the comprehensive molecular and genetic tumour makeup, may overcome these challenges and allow for the development of tailored treatment plans. By integrating genomic data and other biomarkers, precision radiotherapy has the potential to maximise tumour control while minimising toxicity to surrounding healthy tissues.
Tumour response
Technological advancements in radiotherapy have increased the potential of physical radiation tailoring to personalise treatment. However, the optimisation process typically focuses on dose conformality, ignoring biological factors and assuming that all tumours react similarly to radiation (Price et al., Reference Price, Prabhakaran and West2023). Unlike medical oncology, where genomic signatures have become part of routine practice (e.g., MammaPrint tests, Oncotype DX or PAM50), their use in radiotherapy has been limited (Parker et al., Reference Parker, Hunter, Ghazi, Hayeems, Rousseau and Miller2023). Meanwhile, radiation impacts several molecular pathways, such as DNA damage, hypoxia or proliferation (Reisz et al., Reference Reisz, Bansal, Qian, Zhao and Furdui2014; Wang et al., Reference Wang, Wang and Qian2018; Huang and Zhou, Reference Huang and Zhou2020).
Several somatic mutations have already been established as conferring radioresistance. Numerous studies, including breast (Jameel et al., Reference Jameel, Rao, Cawkwell and Drew2004), colorectal (Munro et al., Reference Munro, Lain and Lane2005) and head and neck cancers (Hutchinson et al., Reference Hutchinson, Mierzwa and D’Silva2020), gliomas (Werbrouck et al., Reference Werbrouck, Evangelista, Lobón-Iglesias, Barret, Le Teuff, Merlevede, Brusini, Kergrohen, Mondini, Bolle, Varlet, Beccaria, Boddaert, Puget, Grill, M-A and Castel2019) and sarcomas (Casey et al., Reference Casey, Pitter, Wexler, Slotkin, Gupta and Wolden2021) have shown that TP53 mutations might impair radiotherapy response. Other notable examples are KEAP1 and NFE2L2/NRF2 mutations in NSCLC and head and neck cancers (Binkley et al., Reference Binkley, Jeon, Nesselbush, Moding, Nabet, Almanza, Kunder, Stehr, Yoo, Rhee, Xiang, Chabon, Hamilton, Kurtz, Gojenola, Owen, Ko, Shin, Maxim, Lui, Backhus, Berry, Shrager, Ramchandran, Padda, Das, Neal, Wakelee, Alizadeh, Loo and Diehn2020; Guan et al., Reference Guan, Nambiar, Cao, Viswanathan, Kwok, Hui, Hou, Hildebrand, von Eyben, Holmes, Zhao, Kong, Wamsley, Zhang, Major, Seol, Sunwoo, Hayes, Diehn and Le2023). In addition, the coexistence of KRAS and SMAD4 mutations is an indicator of radioresistance in cervical cancer (Oike et al., Reference Oike, Sekiguchi, Yoshimoto, Takahiro, Ando, Gu, Sasaki, Tokino, Iwase and Ohno2021).
To date, there has been scarce data on molecular predictive signatures in radiotherapy. These examples comprise the PORTOS classifier encompassing 24 genes to predict the efficacy of postoperative radiotherapy in prostate cancer (Zhao et al., Reference Zhao, Chang, Spratt, Erho, Yu, Ashab, Alshalalfa, Speers, Tomlins, Davicioni, Dicker, Carroll, Cooperberg, Freedland, Karnes, Ross, Schaeffer, Den, Nguyen and Feng2016) and the Adjuvant Radiotherapy Intensification Classifier (ARTIC) and POLAR classifiers, which incorporate 27 and 16 genes, respectively, to predict outcomes of postoperative radiotherapy in breast cancer (Sjöström et al., Reference Sjöström, Chang, Fishbane, Davicioni, Zhao, Hartman, Holmberg, Feng, Speers, Pierce, Malmström, Fernö and Karlsson2019, Reference Sjöström, Fyles, Liu, McCready, Shi, Rey-McIntyre, Chang, Feng, Speers, Pierce, Holmberg, Fernö, Malmström and Karlsson2023).
Unlike PORTOS or POLAR, which relate to specific cancers and clinical situations, a radiosensitivity index (RSI) has also been proposed as a pan-cancer and specific marker of cellular radiosensitivity. This index is based on the expression of 10 genes (AR, c-JUN, STAT1, PKC, Rel A, cABL, SUMO1, CDK1, HDAC1 and IRF1) related to DNA damage response, cell cycle, apoptosis and proliferation (Eschrich et al., Reference Eschrich, Pramana, Zhang, Zhao, Boulware, Lee, Bloom, Rocha-Lima, Kelley, Calvin, Yeatman, Begg and Torres-Roca2009). Based on RSI, a quantitative metric for the biological effect of RT, the genomic-adjusted radiation dose (GARD) has been developed. GARD was initially validated in patients with breast cancer, lung cancer, pancreatic cancer and glioblastoma (Scott et al., Reference Scott, Berglund, Schell, Mihaylov, Fulp, Yue, Welsh, Caudell, Ahmed, Strom, Mellon, Venkat, Johnstone, Foekens, Lee, Moros, Dalton, Eschrich, McLeod, Harrison and Torres-Roca2017). This signature was further tested in a pooled, retrospective, pan-cancer cohort and reported as a continuous variable associated with time to first recurrence and overall survival (Scott et al., Reference Scott, Geoffrey, Ellsworth, Scarborough, Ahmed, Oliver, Eschrich, Kattan and Torres-Roca2021). Recently, GARD has been employed in a provocative in silico analysis to explain the unexpected results of the seminal RTOG 0617 trial (unsuccessful radiotherapy dose escalation in locally advanced NSCLC) (Scott et al., Reference Scott, Geoff, Scarborough, Kattan, Peacock, Grass, Mellon, Thapa, Schell, Waller, Poppen, Andl, Teer, Eschrich, Dilling, Dalton, Harrison, Fox and Torres-Roca2021). The authors assumed that this model allows for deriving an optimal radiation dose in each patient. Another study employing prospectively collected tissues showed that low RSI values (denoting higher radiosensitivity) are associated with increased immune infiltration and activation (Grass et al., Reference Grass, Alfonso, Welsh, Ahmed, Teer, Pilon-Thomas, Harrison, Cleveland, Mulé, Eschrich, Enderling and Torres-Roca2022). Recently, based on the reanalysis of the publicly available datasets – Merged Microarray-Acquired Dataset (Bin Lim et al., Reference Bin Lim, Chua, Yeong, Tan, W-T and Lim2019) and the Cancer Genome Atlas (Weinstein et al., Reference Weinstein, Collisson, Mills, Shaw, Ozenberger, Ellrott, Shmulevich, Sander and Stuart2013) – RSI was shown to be associated with immune-related features and predicted response to PD-1 blockade (Dai et al., Reference Dai, Wang, Shen, Lo, Yang, Lin, H-L and Huang2021). However, a recent analysis showed that RSI is not associated with survival and should not be used for radiation dose adjustments (Mistry, Reference Mistry2023). It was also suggested that the RSI of tumour clones remaining after RT, instead of the initial tumour population, should be evaluated to better predict the RT outcome (Alfonso and Berk, Reference Alfonso and Berk2019).
Incorporating genomic signatures in radiotherapy decision-making has shown significant advancement through recent research, such as the GARD-based trial, to optimise radiotherapy for triple-negative breast cancer (NCT05528133). The European Organisation for Research and Treatment of Cancer has appraised the evidence from RSI/GARD studies as a priority for phase 3 clinical trials in radiotherapy (Thomas et al., Reference Thomas, Eisenhauer, Bristow, Grau, Hurkmans, Ost, Guckenberger, Deutsch, Lacombe and Weber2020). However, the clinical utility of these approaches warrants an evaluation that integrates molecular data into prospective clinical trials and routine clinical practice (Table 2).
Radiotherapy tolerance
The impact of genetic heterogeneity on normal tissue toxicity following radiotherapy is a significant concern in cancer treatment. Individual genetic variations can influence the severity of radiation-induced side effects (Barnett et al., Reference Barnett, West, Dunning, Elliott, Coles, Pharoah and Burnet2009). Normal tissue complications can range from mild to severe and may include skin reactions, inflammation, fibrosis and organ dysfunction (Bentzen, Reference Bentzen2006). However, except for several radiosensitivity syndromes related to biallelic pathogenic mutations in DNA repair genes and deleterious heterozygous ATM mutations in young patients, no genomics-guided radiotherapy is currently used (Bergom et al., Reference Bergom, West, Higginson, Abazeed, Arun, Bentzen, Bernstein, Evans, Gerber, Kerns, Keen, Litton, Reiner, Riaz, Rosenstein, Sawakuchi, Shaitelman, Powell and Woodward2019).
Since the beginning of the twenty-first century, more than 100 articles analysing the impact of DNA sequence changes on the frequency and severity of radiation-induced complications have been published (Andreassen et al., Reference Andreassen, Schack, Laursen and Alsner2016). Most of these studies have addressed SNVs, which typically affect the genes responsible for processes such as DNA break or inflammation. However, these studies were usually small (median of approximately 150 patients), hence lowering the statistical power for comparisons (Andreassen et al., Reference Andreassen, Schack, Laursen and Alsner2016).
To reduce the bias associated with the publication of numerous low-quality studies, the Radiogenomics Consortium (https://epi.grants.cancer.gov/radiogenomics/) was created in 2009 (West et al., Reference West, Rosenstein, Alsner, Azria, Barnett, Begg, Bentzen, Burnet, Chang-Claude, Chuang, Coles, De Ruyck, De Ruysscher, Dunning, Elliott, Fachal, Hall, Haustermans, Herskind, Hoelscher, Imai, Iwakawa, Jones, Kulich, Langendijk, O’Neils, Ozsahin, Parliament, Polanski, Rosenstein, Seminara, Symonds, Talbot, Thierens, Vega, Catherine and Yarnold2010). This initiative allowed for assembling adequate groups of patients with diverse clinical characteristics and validating presumed associations of SNVs with radiation toxicity. However, the results of the prospective study published in 2012 were a huge disappointment because none of the reported relationships (98 SNVs in 46 genes) were confirmed (Barnett et al., Reference Barnett, Coles, Elliott, Baynes, Luccarini, Conroy, Wilkinson, Tyrer, Misra, Platte, Gulliford, Sydes, Hall, Bentzen, Dearnaley, Burnet, Pharoah, Dunning and West2012). However, this experience prompted the development of research employing large-scale techniques such as genome-wide association studies (GWAS). As a result, potentially interesting SNV associations with radiation reactions were found, such as variants at the locus of the TANC1 gene that was found to be encoding a protein responsible for muscle cell regeneration (Fachal et al., Reference Fachal, Gómez-Caamaño, Barnett, Peleteiro, Carballo, Calvo-Crespo, Kerns, Sánchez-García, Lobato-Busto, Dorling, Elliott, Dearnaley, Sydes, Hall, Burnet, Carracedo, Rosenstein, West, Dunning and Vega2014). The strength of these associations is much higher, with odds ratios of 1.3–1.5, compared with 1.1–1.2 observed in typical GWAS studies (Zhong and Prentice, Reference Zhong and Prentice2010). The Radiogenomics Consortium remains active, and a significant increase in sample size has led to the discovery of several potentially relevant relationships. An analysis of breast and prostate cancer patients from 17 cohorts indicated that the ATM rs1801516 SNP is associated with an increased risk of radiation toxicity (Andreassen et al., Reference Andreassen, Rosenstein, Kerns, Ostrer, De Ruysscher, Cesaretti, Barnett, Dunning, Dorling, West, Burnet, Elliott, Coles, Hall, Fachal, Vega, Gómez-Caamaño, Talbot, Symonds, De Ruyck, Thierens, Ost, Chang-Claude, Seibold, Popanda, Overgaard, Dearnaley, Sydes, Azria, Koch, Parliament, Blackshaw, Sia, Fuentes-Raspall, Cajal, Barnadas, Vesprini, Gutiérrez-Enríquez, Mollà, Díez, Yarnold, Overgaard, Bentzen and Alsner2016). A recent study has revealed a strong association between radiation-induced mucositis and the rs1131769*C locus in the STING1 gene on chromosome 5 (Schack et al., Reference Schack, Naderi, Fachal, Dorling, Luccarini, Dunning, Ong, Chua, Langendijk, Alizadeh, Overgaard, Eriksen, Andreassen and Alsner2022).
A definitive answer to the SNVs’ role in healthy tissues’ response to radiation may come from the international, multicentre REQUITE project (www.requite.eu) funded by the European Union through its 7th Framework Programme (West et al., Reference West, Azria, Chang-Claude, Davidson, Lambin, Rosenstein, De Ruysscher, Talbot, Thierens, Valdagni, Vega and Yuille2014). The project, performed in collaboration with the Radiogenomics Consortium, aimed at predicting and reducing the risk of long-term side effects of radiotherapy and completed patient recruitment (Seibold et al., Reference Seibold, Webb, Aguado-Barrera, Azria, Bourgier, Brengues, Briers, Bultijnck, Calvo-Crespo, Carballo, Choudhury, Cicchetti, Claßen, Delmastro, Dunning, Elliott, Fachal, Farcy-Jacquet, Gabriele, Garibaldi, Gómez-Caamaño, Gutiérrez-Enríquez, Higginson, Johnson, Lobato-Busto, Mollà, Müller, Payne, Peleteiro, Post, Rancati, Rattay, Reyes, Rosenstein, De Ruysscher, De Santis, Schäfer, Schnabel, Sperk, Symonds, Stobart, Taboada-Valladares, Talbot, Valdagni, Vega, Veldeman, Ward, Weißenberger, West and Chang-Claude2019). REQUITE reported that polygenic risk scores (PRS) may be clinically useful and that incorporation of SNP-SNP interactions improves patient classification and prediction of radiotherapy-related toxicity (Franco et al., Reference Franco, Massi, Ieva, Manzoni, Paganoni, Zunino, Veldeman, Ost, Fonteyne, Talbot, Rattay, Webb, Johnson, Lambrecht, Haustermans, De Meerleer, de Ruysscher, Vanneste, Van Limbergen, Choudhury, Elliott, Sperk, Veldwijk, Herskind, Avuzzi, Noris Chiorda, Valdagni, Azria, Farcy-Jacquet, Brengues, Rosenstein, Stock, Vega, Aguado-Barrera, Sosa-Fajardo, Dunning, Fachal, Kerns, Payne, Chang-Claude, Seibold, West and Rancati2021). This project significantly advanced the collaborations among stakeholders, including healthcare professionals, researchers and industry partners, highlighting the importance of personalised radiotherapy. Other collaborative genetic association studies at both the national and global levels include Gene-PARE (Ho et al., Reference Ho, Atencio, Peters, Stock, Formenti, Cesaretti, Green, Haffty, Drumea, Leitzin, Kuten, Azria, Ozsahin, Overgaard, Andreassen, Trop, Park and Rosenstein2006), RadGenomics (Iwakawa et al., Reference Iwakawa, Imai, Harada, Ban, Michikawa, Saegusa, Sagara, Tsuji, Noda and Ishikawa2002) and RAPPER (Burnet et al., Reference Burnet, Barnett, Elliott, Dearnaley, Pharoah, Dunning and West2013). Understanding the impact of radiogenomic heterogeneity on normal tissue radiation toxicity is essential for developing more effective and safe personalised strategies.
In summary, for a long time, radiotherapy optimisation was focused on dosage conformity rather than biological factors. It is critical to factor in tumour heterogeneity when considering radiotherapy outcomes; hence, developing and verifying molecular signatures such as PORTOS, ARTIC and POLAR, together with pan-cancer RSI, constitute the base for resolving this predicament.
Other factors
Circadian heterogeneity
The efficacy of anticancer treatment may also be affected by circadian rhythm, a biological phenomenon displaying endogenous, untrainable 24-h oscillation (Lee, Reference Lee2021). The circadian clock regulates several key processes in the human body, including metabolism and cell division, with 40% of the transcriptome under circadian control in at least some tissues (Ruben et al., Reference Ruben, Wu, Smith, Schmidt, Francey, Lee, Anafi and Hogenesch2018). Recently, a comprehensive analysis of clock genes across different human cancers was performed using primary solid tumour data from The Cancer Genome Atlas (Ye et al., Reference Ye, Xiang, Ozguc, Kim, Liu, Park, Hu, Diao, Lou, Lin, Guo, Zhou, Wang, Chen, Takahashi, Mills, S-H and Han2018). Based on the available evidence, the International Agency for Research on Cancer classified shift work that involves circadian disruption as potentially carcinogenic to humans (IARC Monographs Vol 124 group, 2019).
Chronotherapy involves administering treatment at specific times of day to optimise its effectiveness and minimise side effects (Zhou et al., Reference Zhou, Wang, Zhang and Tang2021). This approach has been tested in several clinical trials with conflicting results: some showed improved efficacy and reduced toxicity (Lévi et al., Reference Lévi, Zidani and Misset1997; Giacchetti et al., Reference Giacchetti, Bjarnason, Garufi, Genet, Iacobelli, Tampellini, Smaaland, Focan, Coudert, Humblet, Canon, Adenis, Lo Re, Carvalho, Schueller, Anciaux, Lentz, Baron, Gorlia and Lévi2006), whereas others did not demonstrate significant differences (Garufi et al., Reference Garufi, Vanni, Aschelter, Zappalà, Bria, Nisticò, Sperduti, Cognetti and Terzoli2006; Qvortrup et al., Reference Qvortrup, Jensen, Fokstuen, Nielsen, Keldsen, Glimelius, Bjerregaard, Mejer, Larsen and Pfeiffer2010). Some data indicate that chronomodulation might be relevant in the context of immunotherapy. For instance, a recent provocative study reported inferior overall survival in patients who received more than 20% of immunotherapy infusions after 4:30 PM (Qian et al., Reference Qian, Kleber, Brammer, Xu, Switchenko, Janopaul-Naylor, Zhong, Yushak, Harvey, Paulos, Lawson, Khan, Kudchadkar and Buchwald2021). However, these observations warrant verification in prospective randomised clinical trials. Data pertinent to radiotherapy comes from the REQUITE project, which disclosed novel serendipitous associations, for example, the interaction between time, circadian rhythm-related genes (CLOCK, PER3 and RASD1) and late radiation toxicity in breast cancer patients (Webb et al., Reference Webb, Harper, Rattay, Aguado-Barrera, Azria, Bourgier, Brengues, Briers, Bultijnck, Chang-Claude, Choudhury, Cicchetti, De Ruysscher, De Santis, Dunning, Elliott, Fachal, Gómez-Caamaño, Gutiérrez-Enríquez, Johnson, Lobato-Busto, Kerns, Post, Rancati, Reyes, Rosenstein, Seibold, Seoane, Sosa-Fajardo, Sperk, Taboada-Valladares, Valdagni, Vega, Veldeman, Ward, West, Symonds and Talbot2022).
Microbiome heterogeneity
The human microbiome, comprising trillions of diverse microbial organisms, plays a significant role in modulating health and disease states, including cancer (Hou et al., Reference Hou, Wu, Chen, Wang, Zhang, Xiao, Zhu, Koya, Wei, Li and Chen2022). Recent research indicates that microbiome heterogeneity can greatly influence response to anticancer treatment via drug-microbiota interactions.
Bacteria-derived enzymes target chemical compounds, including drugs used in systemic treatment. For example, approximately 40% of patients treated with irinotecan experience severe mucositis, sometimes leading to treatment cessation. Irinotecan is converted into its active form, SN38, which is later reverted back to an inactive form, SN38G, in the liver. Bacterial β-glucuronidases can then convert SN38G in the gastrointestinal tract back to its toxic form (Wallace et al., Reference Wallace, Wang, Lane, Scott, Orans, Koo, Venkatesh, Jobin, Yeh, Mani and Redinbo2010). Additionally, Bifidobacterium longum, Collinsella aerofaciens and Enterococcus faecium abundancy in stool was found to be associated with increased response to anti–PD-1 treatment in patients with melanoma (Matson et al., Reference Matson, Fessler, Bao, Chongsuwat, Zha, Alegre, Luke and Gajewski2018).
Additionally, the gut microbiome generates numerous metabolites, with short-chain fatty acids (SCFAs) being among the most prevalent and crucial. Significantly, SCFAs act as secondary messengers that facilitate signal transmission and influence the onset and progression of various diseases. Radiotherapy can modify the populations of bacteria that produce SCFAs, leading to changes in SCFA levels, which are linked to several conditions, including radiation-induced intestinal injury (Li et al., Reference Li, Zhang, Wei, He, Ding, Hua, Zhou, Niu, Zhou, Shi, Zhang and Liu2021).
The microbiome studies shape oncologic outcomes and are now being leveraged for the development of novel personalised therapeutic approaches in anticancer treatment. However, this topic exceeds the scope of this paper and has been addressed elsewhere (Chrysostomou et al., Reference Chrysostomou, Roberts, Marchesi and Kinross2023; Yi et al., Reference Yi, Lu, Shen, Wu and Zhang2023).
Conclusions
Precision medicine has made remarkable progress in oncology by promising to administer therapy to “the right patient at the right time” (Abrahams, 2008). This review has discussed the impact of cancer heterogeneity as a major challenge facing precision oncology development. Apart from affecting treatment outcomes, heterogeneity can also be employed in the context of prevention and early detection. The results of the first large-scale observational cohort study evaluating methylation-based multicancer early detection diagnostic test (SIMPLIFY) have demonstrated the feasibility of this approach (Rebbeck et al., Reference Rebbeck, Burns-White, Chan, Emmons, Freedman, Hunter, Kraft, Laden, Mucci, Parmigiani, Schrag, Syngal, Tamimi, Viswanath, Yurgelun and Garber2018; Nicholson et al., Reference Nicholson, Oke, Virdee, Harris, O’Doherty, Park, Hamady, Sehgal, Millar, Medley, Tonner, Vargova, Engonidou, Riahi, Luan, Hiom, Kumar, Nandani, Kurtzman, Yu, Freestone, Pearson, Hobbs, Perera and Middleton2023; Tie, Reference Tie2023).
Genomic makeup has been shown to impact the effectiveness and toxicity of systemic treatments and radiotherapy. Thus, genomic testing can identify pathogenic gene variants and polymorphisms affecting drug metabolism or mechanism of action, thus increasing the risk of treatment failure or toxicity. Spatial and temporal tumoural heterogeneity is a complex phenomenon linked to resistance to therapy, disease progression and adverse prognosis. There are substantial genetic and molecular differences across various tumour regions and between primary and metastatic foci. A better understanding of this phenomenon allowed for the development of novel strategies, for example, targeting with systemic therapies or radiation-specific tumour regions or populations of cancer cells.
Implementing advanced technologies, such as NGS, liquid biopsies and imaging modalities, has fostered precision oncology, accounting for both genomic and tumoural heterogeneity. NGS is routinely used to examine mutations in, for example, EGFR, BRAF and ALK, which are molecular targets for modern therapies. Liquid biopsies, which involve analysing circulating tumour cells or circulating tumour DNA, offer a noninvasive way to identify genetic alterations and monitor tumour progression. Several clinical trials, including the ongoing NCI-MATCH and MASTER trials and the previously completed MyPathway and MPACT trials, have been designed to identify genetic mutations associated with specific targeted therapies and develop novel treatment strategies for overcoming therapy resistance. These trials are expected to prompt further development of precision oncology.
As precision oncology continues to evolve, the future holds great promise for overcoming current challenges. Advanced tools should be more accessible and affordable. There is a need for routine, more comprehensive genomic profiling of different regions of primary and metastatic tumours to fully understand cancer heterogeneity. In addition, integrating machine learning algorithms and artificial intelligence would allow better identification of new therapeutic targets and the development of even more personalised treatment strategies. An exciting area of future research in precision oncology is the use of combination therapies that simultaneously target multiple pathways and molecular targets; this approach has the potential to overcome heterogeneity-led resistance to single-agent targeted therapies. Another area for improvement is integrating precision oncology into clinical practice and expanding access to new technologies for community oncologists and patients. This will require the development of user-friendly platforms and tools that are easily integrable into clinical workflows.
Overall, precision oncology holds great promise for improving cancer treatment efficacy by enabling personalised treatment strategies based on unique cancer and patient characteristics. Although challenges remain to be addressed, ongoing research and emerging developments create real hope for breath-taking therapeutic approaches and improved patient outcomes.
Open peer review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/pcm.2023.23.
Author contribution
J.J. and B.T. developed the paper concept and outline. B.T., F.G., M.Br. and M.Bi. collected and assembled the data. J.J. provided administrative support and oversight and revised the text. F.G. prepared the figures. B.T., F.G., M.Br., M.Bi. and J.J. wrote and approved the manuscript. B.T. and F.G. contributed equally to this work.
Financial support
F.G. is supported by Polish National Science Centre Grants PRELUDIUM 2018/31/N/NZ5/03214, ETIUDA 2020/36/T/NZ5/00610, and Polish Stem Cells Bank grant ExCELLent.
Competing interest
J.J. declares advisory roles for BMS, Roche and MSD, lectures for Roche (not compensated), Pfizer, Novartis and MSD and travel support from Takeda. Other authors have no conflicts of interest to declare.







Comments
Gdańsk, 9 May, 2023
Professor Dame Anna Dominiczak
Editor-in-Chief Cambridge Prisms: Precision Medicine
Dear Prof. Dominiczak,
On behalf of all authors, I am pleased to submit solicited manuscript entitled “Diversity in precision oncology”.
Precision oncology is a rapidly evolving concept with great promise in cancer treatment. However, a complex cancer diversity attributed to genomic and acquired tumour heterogeneity limits treatment effectiveness and increases toxicity. By understanding cancer diversity and utilising advanced tools to personalise treatment strategies, precision oncology has the potential to revolutionise cancer care. In this article, we review the current status of precision oncology, specifically focusing on the impact of tumour diversity and genomic patient features on systemic therapies and radiation. We also discuss implementing novel tools, such as next-generation sequencing and liquid biopsies, to overcome this problem.
We humbly thank you for your time in considering our manuscript for publication. Please let us know if you have any further comments or concerns.
We look forward to hearing from you.
Yours faithfully,
Prof. Jacek Jassem, MD PhD