1. Introduction
A common problem in biomedical research is low reproductive performance of genetically modified model mice, which in turn has a negative impact on the production of new knowledge in medicine. However, some strains of mice have, after many years of selection, become more resistant to different types of environmental stress and produce larger litters in most types of animal facilities. Such strains are often denoted ‘high breeders’, and they carry genes ensuring large litter size, high stress resistance during pregnancy and good nursing properties during the lactation period. The NFR/N strain (an inbred NMRI-derived strain) is an example of an inbred strain with high breeding performance, while common C57BL strains normally produce lower number of liters, and are often denoted ‘moderate breeders’. Females of the NFR/N strain are known to produce several large litters during a long period of time. Furthermore, they are known to be excellent mothers exhibiting very good nursing properties. Therefore, it is likely that NFR/N mice carry genetic polymorphisms (several modified genes) that are of importance for successful reproduction in mice.
By studying the second backcross (N2) of the inbred mouse strains NFR/N and C57B1/10.Q (denoted B10.Q), we previously succeeded to identify a number of new quantitative trait loci (QTLs) involved in the maternal control of pregnancy success (Liljander et al., Reference Liljander, Sällström, Andersson, Wernhoff, Andersson, Holmdahl and Mattsson2006). The loci identified are now denoted Pregq1, Pregq2, Pregq3 and Pregq4 affecting pregnancy rate; Fecq3 and Fecq4 controlling fecundity (litter size); and Neogq1, which is a maternal locus that is affecting the growth of the pups during the early lactation period.
The present paper is focusing exclusively on the Fecq4 locus at chromosome 9 that seems to contain polymorphic genes that positively can affect the fecundity (litter size) in female mice. We find locus particularly interesting since, in addition, other groups have found QTLs of potential interest for both litter size (Kirkpatrick et al., Reference Kirkpatrick, Mangelt, Schulman and Martin1998) developments of primary oocytes (Everett et al., Reference Everett, Auchincloss, Kaufman, Abbott and West2004) and super ovulation rate (Spearow & Barkley, Reference Spearow and Barkley1999; Spearow et al., Reference Spearow, Doemeny, Sera, Leffler and Barkley1999a, Reference Spearow, Nutson, Mailliard, Porter and Barkleyb) at this region of chromosome 9. The QTL observed by Kirkpatrick et al. (Reference Kirkpatrick, Mangelt, Schulman and Martin1998) was of suggestive significant, while the QTLs for primary oocyte development and super-ovulation rate both were significant. Mice different from C57B1 and NMRI had used in these studies. This convinced us that the Fecq4 locus contains one or more polymorphic genes that are of general importance for the control of reproductive performance in female mice.
In the present paper, we provide data from B10.Q.NFR/N-Fecq4 congenic mice (Fecq4 locus of NFR/N origin in B10.Q mice, from now only denoted Fecq4 congenics) showing that both litter size, super-ovulation rate and embryo development in vitro is significantly elevated in Fecq4 congenic compared with wild-type B10.Q female mice. We also discuss the possible role of candidate genes within this region of chromosome 9.
2. Material and methods
(i) Animals
NFR/N mice were originally obtained from the National Institute of Health (Maryland, USA) and the B10.Q mice were bought form The Jackson Laboratory (Bar Harbor, Maine, USA). (B10.Q ∝ NFR/N) ∝ B10.Q N8 mice were bred in IVC-cages in the BMC barrier animal house and at the Biomedical Center and the Pathology animal house at Lund University, Sweden. All mice were fed ad libitum with standard rodent pellets (LAB FOR R36, irradiated breeding food for rats and mice, Lactamin AB, Sweden) and water in a climate-controlled environment with a photoperiod of LD 12:12. The mice used in the present study had clean health monitoring protocols according to the Federation of European Laboratory Animal Sciences Association (FELASA) recommendations. Ethical permissions: M125-04 (embryo transfer) and M236-06 (reproduction and arthritis).
(ii) Description of breeding cages
A total number of 17 Fecq4 females and 20 control B10.Q females were used in the study of litter size. The set-up of the breeding cages were as follows: two females were kept together with one B10.Q male for five month. During this time the number of litters and pups were counted. All the females were 10 weeks old when the study started.
(iii) Description of the congenic strain and the Fecq4 locus
The B10.Q.Fecq4 congenic mice were produced by traditional back-crossing through the replacement of a selected region from the NFR/N on chromosome 9. Briefly, mice from an NFR/N ∝ B10.Q N2 backcross, heterozygous for selected markers on chromosome 9, were backcrossed to B10.Q for eight generations (N8). At this stage the mice are considered to be 99·6% B10.Q homozygous on all the other chromosomes. Mice heterozygous for markers between D9Mit27 (50·4 Mb) and D9mit124 (75·9 Mb) were then intercrossed two times to produce the congenic line Fecq4. All the mice in the study are homozygous and the fragment size is equal in all animals (Fig. 1). To exclude the possibility that there may have been any NFR/N alleles selectively retained during backcrossing at other known QTLs for reproductive loci, we genotyped the following loci: Fecq1 and Fecq2 (Kirkpatrick et al., Reference Kirkpatrick, Mangelt, Schulman and Martin1998), Fecq3, Pregq1, Cia40/Pregq2, Pregq3 and Pregq4 (Liljander et al., Reference Liljander, Sällström, Andersson, Wernhoff, Andersson, Holmdahl and Mattsson2006a, Reference Liljander, Sallstrom, Andersson, Andersson, Holmdahl and Mattssonb; Reference Liljander, Andersson, Holmdahl and Mattsson2008). The genotype analysis showed that no remaining NFR/N fragments were present at these loci.

Fig. 1. The dark area indicates the NFR/N congenic fragment, Fecq4 in the congenic mice B10.Q.NFR/N-Fecq4. The markers are placed according to Mouse Ensemble built 36 (http://www.ensembl.org/Mus_musculus/index.html).
(iv) Super-ovulation and embryo preparation
The standard protocols for super-ovulation and embryo preparation from oviducts (Nagy et al., Reference Nagy, Gertsenstein, Vintersten and Behringer2003) were followed. Briefly, age matched, 4 weeks old female mice were first injected i.p. with 5 i.u. of the follicle stimulating hormone PMSG (G-4877, SIGMA) at 2 p.m. in the afternoon. At noon, 46 h later, the females received an i.p. injection of 5 i.u of the ovulation-inducing hormone hCG (C-1063, SIGMA) and were placed together with single-caged adult B10.Q males (1 female/male). Mating normally takes place in the middle of the night, and on the following morning (E.D. day 0·5) the females were inspected for vaginal plugs. The females were sacrificed and oviducts removed. Oviducts from each mouse were put in tubes with medium M2 (M7167, SIGMA). The 1-cell embryos were then prepared under a dissection microscope, in drops of M2 medium supplemented with hyaluronidase (makes the embryos separate), and washed several times in medium M2 and before subjected to in vitro cultivation.
(v) In vitro cultivation of pre-implantation embryos
The in vitro cultivation of the pre-implantation embryos was, in principle, performed according to the standard protocol (Nagy et al., Reference Nagy, Gertsenstein, Vintersten and Behringer2003). Briefly, the one-cell embryos were transferred to small Petri dishes containing 30 μl drops of medium M16 (M7292, SIGMA) covered with mineral oil. Each drop contained the total yield of 1-cell embryos from one individual mouse. The embryos were incubated at 37°C in humidified air containing 5% CO2. The number of viable embryos of expected developmental stage (2-cell embryos day 1·5, morula day 2·5 and blastocysts day 3·5 and 4·5) was counted once every day.
(vi) Micro-satellite genotyping
Toe biopsies were collected from all females and DNA was isolated according to a previously described protocol (Laird et al., Reference Laird, Zijderveld, Linders, Rudnicki and Jaenisch1991). The congenic fragment was genotyped with ten fluorescence-labelled micro-satellite markers (INTERACTIVA, Ulm, Germany) in order to verify the size of the congenic fragment. PCR amplification was performed according to a previously described protocol (Laird et al., Reference Laird, Zijderveld, Linders, Rudnicki and Jaenisch1991). The following program was used to amplify the DNA: denaturation at 95°C for 3 min, annealing at 56°C for 45 s, polymerization at 72°C for 1 min, 30 cycles of 95°C for 30 s, 56°C for 45 s, 72°C for 1 min and a final extension step of 7 min at 72°C. The PCR products were analysed on a MegaBACE™ 1000 (Amersham Pharmacia Biotech) according to the manufacture's protocol. Data were analysed with Genetic Profiler 1·5 through comparison from parental mouse strains.
(vii) Statistical analysis
Statistical analyses were performed by the two-tailed Mann–Whitney U-test.
3. Results
(i) Breeding capacity of Fecq4 congenic mice
Fecq4 congenic mice produced significantly more pups per female than homozygous B10.Q littermates when counting the production of pups over a five months period of time (Fig. 2a). Furthermore, the Fecq4 congenic mice also produced significantly more litters (P=0·011) over the same period of time. Although Fecq4 congenic mice characteristically showed a larger litter size (7·6 pups/Fecq4 congenic female compared with 6·3 pups/B10.Q female), this difference was not significant (P=0·094, Fig. 2b).
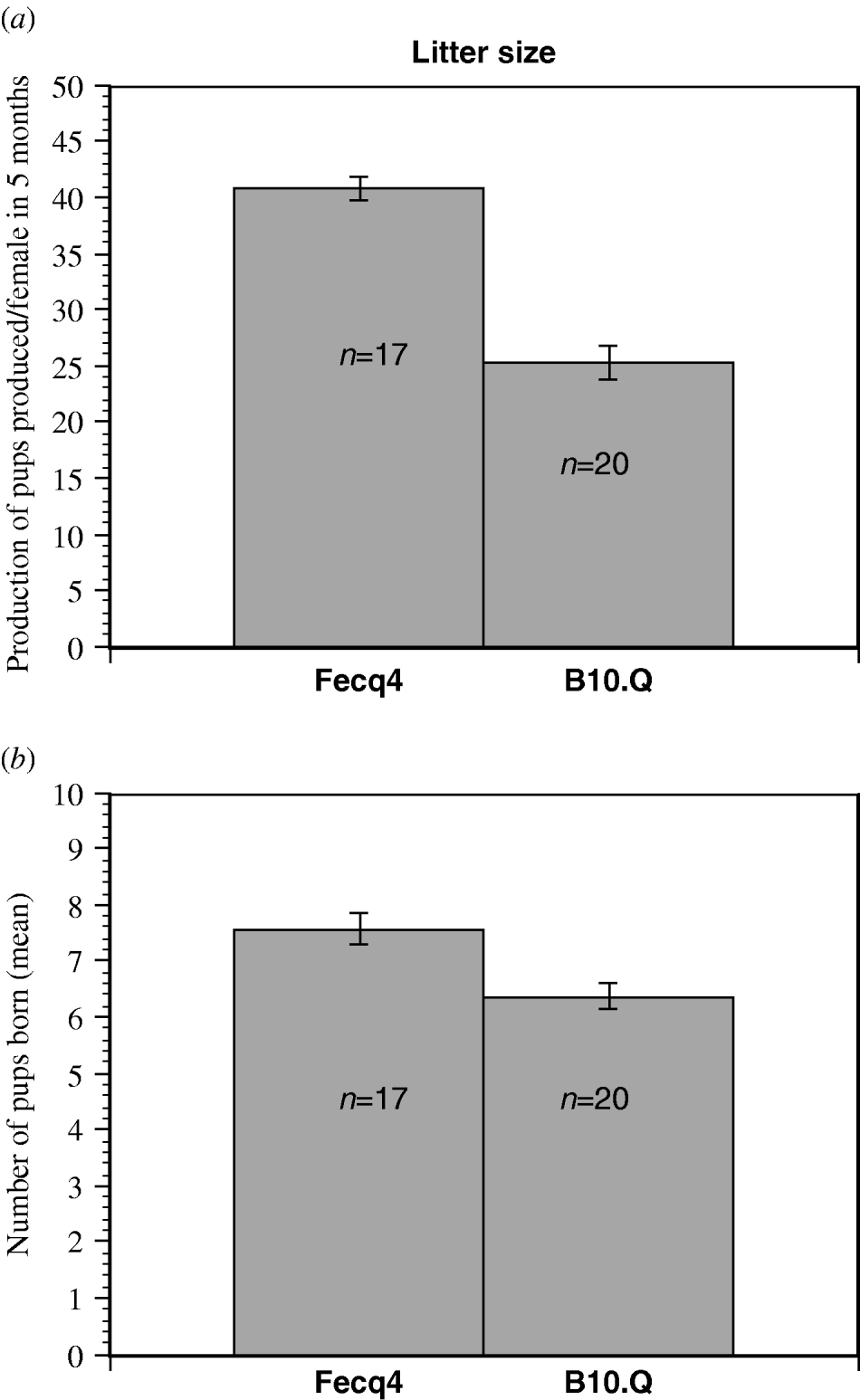
Fig. 2. (a) The total production of pups per female in congenic B10.Q.NFR/N-Fecq4 (Fecq4) and B10.Q control mice over a five months period (mean value S.E). P=0·035. (b) The mean value (S.E.) for the litter size from each female over a five-month of period. Although B10.Q.NFR/N-Fecq4 mice exhibited a higher litter size, the difference was not quite significant (P=0·094).
(ii) Oocyte yield in response to super-ovulation and embryo development in vitro
As outlined in Fig. 3a and b the congenic Fecq4 mice responded more efficiently to a standard super ovulation protocol compared with the B10.Q wt mice. The congenic mice used in the present study were all homozygous for the congenic NFR/N fragment. The congenic and the wild-type mice were all of the same age (4 weeks), and both congenic and wt controls were allowed to mate with adult, fertile B10.Q male mice. The in vitro development of fertilized embryos was more efficient in the embryos that were heterozygous for the Fecq4 fragment. In fact the median egg yield in response to hormone treatment (day 0·5) was doubled, but the development to the blastocyst stage (day 3·5–4·5) was almost 4–5-fold increased. The lower yield of blastocysts on day 3·5 compared with day 4·5 was due to the fact that some embryos still were at the morula stage on day 3·5. Unfertilized negative control embryos (embryos prepared from super ovulated females that were not allowed to go with males) characteristically died by two main pathways: lysis or uncontrolled cell division (Fig. 4).

Fig. 3. (a) Difference in the number of embryos obtained after super-ovulation in B10.Q.NFR/N-Fecq4 (Fecq4) and B10.Q control females. Mean value (S.E.) from ten (n=10) Fecq4 and ten (n=10) B10.Q females at 4 weeks of age. (b) Difference in the yield of embryos after super-ovulation in B10.Q.NFR/N-Fecq4 congenic mice (Fecq4) and B10.Q controls. Mean value (S.E.) from n=34 Fecq4 congenic females and n=37 B10.Q females (P=0·001).

Fig. 4. Common shape of unfertilized B10.Q embryos. The pictures show the most common shapes of unfertilized B10.Q wild-type and B10.Q.NFR/N-Fecq4 congenic mouse embryos after 24 h of in vitro cultivation. Characteristically, the embryos die quickly by lysis (a) or undergo uncontrolled cell division leading to a ‘morula-like’ shape (b). These two ‘death pathways’ of unfertilized embryos were equally common in the strains studied. A small proportion (<5%) of the unfertilized B10.Q wild-type and B10.Q.NFR/N-Fecq4 congenic embryos did undergo parthenogenetic cell division, which is almost impossible to distinguish from ordinary cell division.
Figure 5 shows in vitro development of pre-implantation embryos day 0·5–4·5. The characteristic difference between day 3·5 and day 4·5 blastocysts is obvious. On day 3·5 many morulae have not yet developed the blastocoels, and those blastocysts that are visible are at an early development stage. On day 4·5 almost all embryos has reached the blastocyst stage, and many of them have also hatched (been released from egg shell – the zona pellucida) and expanded.

Fig. 5. Development of embryos day 0·5–4·5.
4. Discussion
A limited number of linkage analyses based on QTL mapping have been performed for identification of gene regions controlling litter size in female mice (Kirkpatrick et al., Reference Kirkpatrick, Mangelt, Schulman and Martin1998; Peripato et al., Reference Peripato, De Brito, Vaughn, Pletscher, Matioli and Cheverud2002, Reference Peripato, De Brito, Matioli, Pletscher, Vaughn and Cheverud2004). As previously mentioned, Kirkpatrick et al. (Reference Kirkpatrick, Mangelt, Schulman and Martin1998) reported a locus of suggestive significance for the trait litter size close to the Fecq4 locus, while Peripato et al. (Reference Peripato, De Brito, Vaughn, Pletscher, Matioli and Cheverud2002, Reference Peripato, De Brito, Matioli, Pletscher, Vaughn and Cheverud2004) did not detect any significant loci for this trait on chromosome 9. However, the fact that Everett et al. (Reference Everett, Auchincloss, Kaufman, Abbott and West2004), Spearow & Barkley (Reference Spearow and Barkley1999) and Spearow et al. (Reference Spearow, Doemeny, Sera, Leffler and Barkley1999a, Reference Spearow, Nutson, Mailliard, Porter and Barkleyb) did detect loci, significantly linked to ovulation traits, in the vicinity of the Fecq4 locus convinced us that this region of chromosome 9 indeed contain polymorphic genes important for optimizing breeding success in female mice. It is possible that polymorphisms or mutations in the same gene/s affect both spontaneous ovulation and artificial hormone-induced ovulation (super ovulation), as well as litter size. For these reasons, we found it important to produce a congenic Fecq4 mouse line that could be investigated for the traits mentioned above, and that could be used for the final identification of the critical gene/s. In addition, the Fecq4 congenic strain could be a useful tool to optimize breeding in model mice of the C57BL strain that exhibit female-associated breeding problems. Congenic Fecq4 mice could also be valuable in transgenic technology, since the C57BL strain is preferable for the production of ‘classical transgenic’ (by pro-nucleus injection) mice, and the response to super ovulation will determine the yield of injectable embryos.
In fact, the Fecq4 mouse strain has already been used at our laboratory to facilitate the freezing and embryo transfer of two-cell embryos from other congenic strains (strains that were undergoing backcrossing against B10.Q, and quickly had to be transferred to another laboratory).
Male mice of two different congenic strains from Dr Holmdahls Laboratory (Medical Inflammation Research, Lund University, Sweden and Karolinska Institute, Department of Medical Biochemistry and Biophysics, Division of Medical Inflammation Research, Sweden), which had to undergo transfer from Lund to Stockholm as frozen embryos, were allowed to mate with super-ovulated, age-matched Fecq4 and B10.Q females. Equal number of Fecq4 and B10.Q females were used for the super-ovulation, but it both cases the final yield of 2-cells were 2–3 times increased if Fecq4 females had been used in mating. The increased yield of frozen embryos obtained by the mating with the Fecq4 females turned out to be critical for the final success of the transfer of these congenic lines. An increased embryo yield from the B6 strain is also very valuable in transgenic technologies – especially when doing pronuclear injections.
The B10.Q mouse is a common mouse in all laboratories working with the H2q-dependent collagen II-induced arthritis models and other H2q-dependent models. Since the ordinary C57BL/6 mouse (the B6 mouse) is a more universal model mouse, we have now started to backcross the Fecq4 congenic strain against B6. We assume that the increase in egg yield by super-ovulation can be of general value in cryopreservation of transgenic or congenic lines on B6 background, or lines that are undergoing backcrossing against B6.
The results of the present study indeed show that the Fecq4 congenic strain has higher breeding performance than ordinary B10.Q wild-type mice, both with respect to litter size and the production of offspring over time. However, even more pronounced is the elevated response to artificial hormone-induced ovulation according to the standard super ovulation protocol, and the dramatically increased yield of viable pre-implantation embryos after in vitro cultivation. The latter property of our Fecq4 congenic line could be used to optimize the result of embryo freezing and embryo transfers of straits that exhibit extremely low response in super ovulation, or are difficult to grow in vitro. Normally, only one or two further back-crossings are needed to remove the Fecq4 fragment.
Although the Fecq4 congenic strain can be useful without further reduction in fragment size, it is of great interest to identify the specific polymorphic or mutated gene(s) that actually are causing the positive effects observed. The Fecq4 fragment contains over 200 genes and a few of these genes have previously been reported to affect reproductive phenotypes that might influence traits such as super-ovulation and litter size. In Table 1, we have listed some of the most interesting candidate genes that are present in the congenic fragment of chromosome 9. We have paid special attention to genes near the linkage peak (58 Mb) of the previously detected QTL (Fecq4), and genes that are known to affect reproductive traits associated with the females.
Table 1. Candidate genes

Summary of possible candidate genes on chromosome 9 for B10.Q.NFR/N-Fecq4 congenic mouse. (http://www.informatics.jax.org/).
Although still very speculative, there are a number of genes that we find particularly interesting. Firstly, genes of the cytochrome P450 family (1, 11 and 19) present in this region all exhibit interesting reproductive phenotypes if mutated (Fisher et al., Reference Fisher, Graves, Parlow and Simpson1998). The Cyp19 gene control the formation of estrogens from C19 steroids, and the process is catalysed by aromatase cytochrome P450 (P450arom). In particular, Cyp19a1 appears to be significant in this context, since null-mutated mice have shown impaired ovulation and impaired reproductive behaviour (mounting). Secondly, a gene of potential interest is the tyrosine kinase gene Chrna3, since null mutations will cause embryonic and postnatal death (Xu et al., Reference Xu, Gelber, Orr-Urtreger, Armstrong, Lewis, Ou, Patrick, Role, De Biasi and Beaudet1999).
Thirdly, the Mpi gene (mannose phosphate isomerase), close to the linkage peak of Fecq4, is also of potential interest, since null-mutated mice exhibit defects in the development of extra-embryonic tissues and embryonic death (DeRossi et al., Reference DeRossi, Bode, Eklund, Zhang, Davis, Westphal, Wang, Borowsky and Freeze2006).
In order to find polymorphisms between NFR/N and B10 mice concerning the genes mentioned above, sequencing analyses are needed. According to known effects of null-mutations, we are paying special attention on the Cyp19a1 gene, and the possible influence of a mutation/polymorphism between the NFR/N and B10.Q strains.
We acknowledge Mary-Ann Sällström and Sara Andersson at Lund Transgenic Core Facility for valuable help with embryo preparations. This study was supported by Österlund's fund, Crafoord's fund, Gustav V 80 year foundation, The Royal Physiographic Society in Lund and The Lars Hierta Memorial Foundation.







