Cheese maturation is a process where microbial fermentation is accompanied by a series of enzymatic and chemical reactions that form the final textural and flavour traits that appeal to the consumer's overall sensorial perception. Even though both artisanal and industrial cheese manufacturing encompass a ripening period, sensorial discrepancies might be found due to the nature of controlled and uncontrolled cheese maturation processes that are carried out (Afshari et al., Reference Afshari, Pillidge, Dias, Osborn and Gill2020b). Probing differences and separating out all parameters that affect ripening have attracted enormous research interest in order to improve cheese production (Madkor et al., Reference Madkor, Tong and El Soda2000; Sgarbi et al., Reference Sgarbi, Lazzi, Tabanelli, Gatti, Neviani and Gardini2013; Gobbetti et al., Reference Gobbetti, De Angelis, Di Cagno, Mancini and Fox2015; Pogačić et al., Reference Pogačić, Maillard, Leclerc, Hervé, Chuat, Yee, Valence and Thierry2015, Reference Pogačić, Maillard, Leclerc, Hervé, Chuat, Valence and Thierry2016; Bancalari et al., Reference Bancalari, Savo Sardaro, Levante, Marseglia, Caligiani, Lazzi, Neviani and Gatti2017; Calasso et al., Reference Calasso, Minervini, De Filippis, Ercolini, De Angelis and Gobbetti2020; Levante et al., Reference Levante, Bancalari, Tambassi, Lazzi, Neviani and Gatti2020; Štefániková et al., Reference Štefániková, Ducková, Miškeje, Kačániová and Čanigová2020). To that end, the complex relationship between microorganisms in cheese and substrates from milk has been investigated to unravel many unknowns of cheese ripening. The ever-increasing amount of information obtained from metabolomics, genomics and proteomics sheds light on the elusive relationship between microbes and substrates within the cheese matrix (Khattab et al., Reference Khattab, Guirguis, Tawfik and Farag2019; Afshari et al., Reference Afshari, Pillidge, Dias, Osborn and Gill2020a). Starter and non-starter lactic acid bacteria (SLAB and NSLAB, respectively) contribute to cheese flavour formation through the assimilation of substrates that are present and/or produced during cheese making. Fermentable sugars in milk, citrate, glycomacropeptides, glycoconjugates of milk-fat globule membranes, N-acetylglucosamine, sialic acid, cell lysate, free amino acids are assimilated from SLAB and NSLAB as carbon sources during the maturation process (Ercan et al., Reference Ercan, Wels, Smid and Kleerebezem2015; Gobbetti et al., Reference Gobbetti, De Angelis, Di Cagno, Mancini and Fox2015; van Mastrigt, et al., Reference van Mastrigt, Gallegos Tejeda, Kristensen, Abee and Smid2018b, Reference van Mastrigt, Egas, Abee and Smid2019; de Møller et al., Reference de Møller, Christensen and Rattray2021). Considering microbial flavour formation by utilization of substrates in milk during cheese ripening, it is crucial to acknowledge two major points: (i) the viability of starter cultures during the ripening and (ii) adequate cell density to form flavour compounds by cellular activity of starter cultures. It was postulated that adjunct culture and/or starter culture can help form flavour compounds during maturation only if they are adequately alive at a certain cell density to perform microbial metabolism through the uptake of substrates in the curd (Nugroho et al., Reference Nugroho, Kleerebezem and Bachmann2020). Since almost all carbon sources are depleted during cheese maturation, the cell viability of SLAB and NSLAB comes into question as does whether dead/alive/injured cells in cheese can still contribute to the flavour formation during ripening (Nugroho et al., Reference Nugroho, Kleerebezem and Bachmann2020). Even though SLAB cell lysis during cheese ripening leads to the release of intracellular enzymes that may catalyse the conversion of amino acids into flavour compounds, it is known that volatile compounds can also be generated through metabolic pathways in which multiple intracellular co-factors must be present. Since these co-factors cannot be supplied by dead cells, prolonged survival of SLAB might more effectively generate volatile compounds. Moreover, flavour-producing enzymes are more prone to rapid denaturation in food matrix compared to those residing in the cell (Wilkinson and LaPointe, Reference Wilkinson and LaPointe2020; Nugroho et al., Reference Nugroho, Kleerebezem and Bachmann2021).
Throughout cheese ripening, microbial utilization of substrates generates cheese taste and aroma. SLAB and NSLAB exploit three major metabolisms for flavour formation: sugar metabolism, proteolysis, and lipolysis (Fig. 1). As a principal carbohydrate for microbial metabolism, lactose is metabolized to lactate, acetate, formate, acetaldehyde and others. In addition to that, citrate-positive lactic acid bacteria (LAB) can produce flavour compounds such as acetoin, 2,3-butanediol, di-acetyl and 2-butanone (Bertuzzi et al., Reference Bertuzzi, McSweeney, Rea and Kilcawley2018b). It is known that proteolytic activity is one of the main triggers to form aroma compounds due to the fact that aromatic amino acids are sources of flavour compounds developed during cheese maturation. As a starting point of proteolysis, proteinases and peptidases from residual coagulant (usually chymosin) and endogenous milk enzymes as well as those from starter and non-starter cultures, degrade milk casein to peptides, which in turn produce free amino acids in the curd as substrates for the subsequent steps. Aminotransferases convert these free amino acids into their corresponding α-keto acids that are further decarboxylated or deaminated into volatile organic compounds (VOCs). When there is a surplus of amino acids within the cell, flavour compounds can be generated by glutamate dehydrogenase in the presence of a receptor, α-ketoglutarate (Smid and Lacroix, Reference Smid and Lacroix2013; Lazzi et al., Reference Lazzi, Povolo, Locci, Bernini, Neviani and Gatti2016; Stefanovic et al., Reference Stefanovic, Kilcawley, Rea, Fitzgerald and McAuliffe2017a). Likewise, flavour compounds can also be produced via lipolysis where milk triglycerides are hydrolysed through lipases and esterases into free fatty acids (FFA), mono- and di-glycerides and glycerol. While short-chain fatty acids directly contribute to flavour, FFAs also serve as precursors to a wide range of other compounds, such as esters, lactones, methyl ketones and secondary alcohols that are associated with diverse cheese flavours (Khattab et al., Reference Khattab, Guirguis, Tawfik and Farag2019).
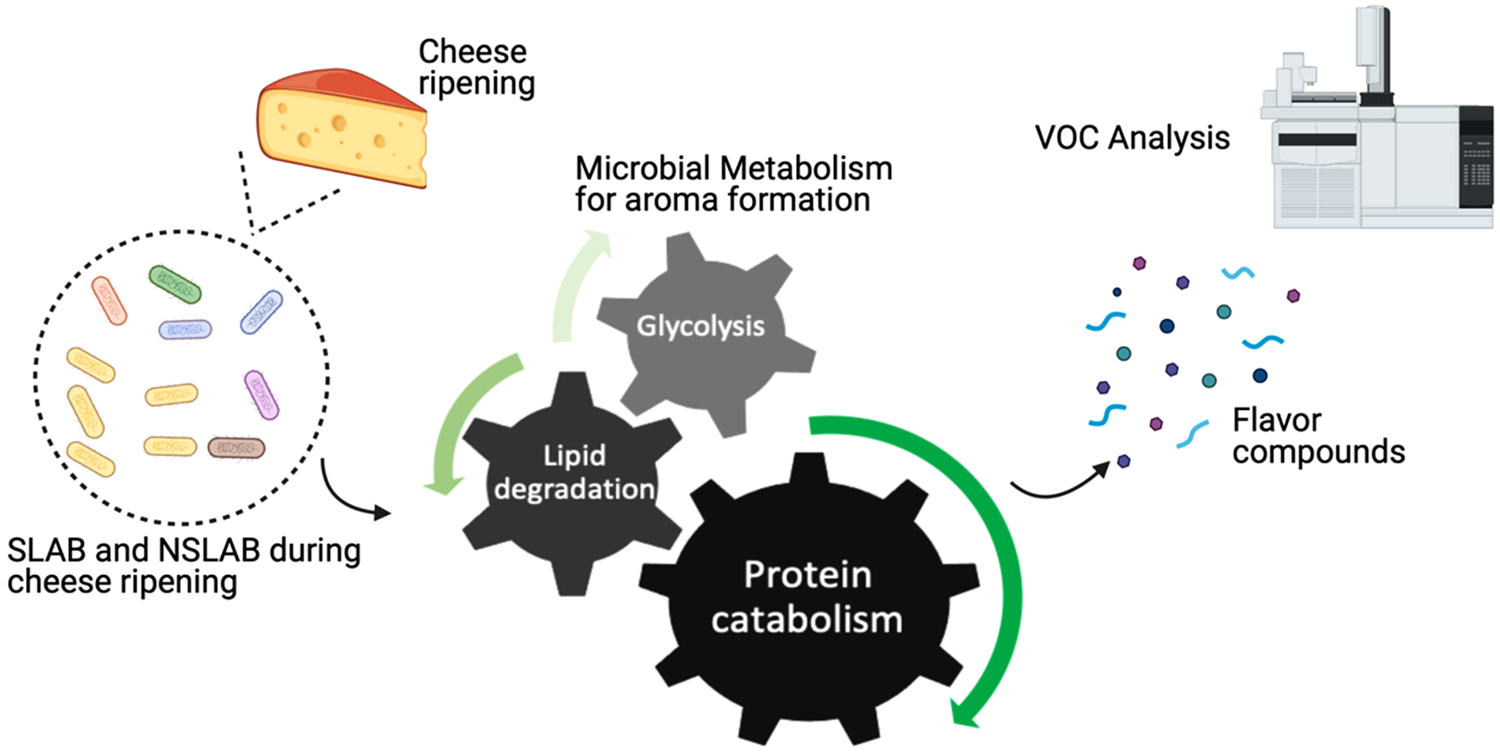
Fig. 1. Schematic representation of aroma formation during cheese ripening.
Acids, alcohols, ketones, sulphur compounds, aldehydes, secondary alcohols, and lactones are among volatile compounds contributing to the final aroma of cheese. Gas chromatography mass spectrometry (GC-MS) has been a prevalent method for monitoring volatile compounds in cheese ripening. The conventional VOC analysis in cheese is a complex procedure mainly comprising of extraction/concentration techniques, gas chromatographic separation of the peaks followed by their identification and quantification (Marilley and Casey, Reference Marilley and Casey2004; Bertuzzi et al., Reference Bertuzzi, McSweeney, Rea and Kilcawley2018b; Terpou et al., Reference Terpou, Bosnea, Kanellaki, Plessas, Bekatorou, Bezirtzoglou and Koutinas2018). Sensitivity and reliability of GC-MS depends on many factors such as the extraction technique, the nature of the extraction polymeric material, the GC column, and the detector used and currently employed extraction methods are quite error-prone, leading to confounding results. Bertuzzi et al. (Reference Bertuzzi, McSweeney, Rea and Kilcawley2018b) recently reviewed the analytical methodologies used for VOC analysis by highlighting the advantages and potential shortcomings of each technique.
In improving cheese quality through the addition of novel non-industrial microbial strains, VOC measurements have highlighted the variation of cheese flavour composition. Hence for adjunct culture selection, alterations in the cheese volatilome due to NSLAB addition were monitored through VOC analysis (Van Hoorde et al., Reference van Hoorde, Van Leuven, Dirinck, Heyndrickx, Coudijzer, Vandamme and Huys2010; Stefanovic et al., Reference Stefanovic, Thierry, Maillard, Bertuzzi, Rea, Fitzgerald, McAuliffe and Kilcawley2017b). Moreover, VOC analysis was also carried out for SLAB with constrained cellular activity (van Mastrigt et al., Reference van Mastrigt, Abee, Lillevang and Smid2018a, Reference van Mastrigt, Gallegos Tejeda, Kristensen, Abee and Smid2018b, Reference van Mastrigt, Egas, Abee and Smid2019) and attenuated NSLAB (Di Cagno et al., Reference Di Cagno, De Pasquale, De Angelis, Buchin, Calasso, Fox and Gobbetti2011) to test their aroma-forming potential (Table 1). With the increasing quest to discover biomarkers of cheese quality, the identification of biomarker compounds detected by GC-MS has gained more attention, especially to determine which compounds might serve as indicators for optimum cheese quality (Afshari et al., Reference Afshari, Pillidge, Dias, Osborn and Gill2021). This research reflection is intended to update recent developments of cheese ripening as determined by volatile profile analysis and to summarize recent efforts in operable aroma formation analysis for the cheese making process.
Table 1. Functional utilization of aroma formation in cheese ripening

VOC analysis to select potential starter cultures for improving cheese flavour
LAB are innate resident microorganisms of cheese. SLAB and NSLAB interactions during cheese ripening may also lead to desired or undesired volatile compounds (van Hoorde et al., Reference van Hoorde, Van Leuven, Dirinck, Heyndrickx, Coudijzer, Vandamme and Huys2010; Sahingil et al., Reference Sahingil, Hayaloglu, Simsek and Ozer2014). Our group investigated whether the metabolic behaviour of a cheese microbial community could be explained by the characterized individual organisms that constituted the culture. To address this question, the dairy-origin LAB Lactococcus lactis subsp. cremoris, Lactococcus lactis subsp. lactis, Streptococcus thermophilus and Leuconostoc mesenteroides, commonly used in cheese starter cultures, were grown in pure culture and as co-cultures. Dynamic metabolic modelling based on the integration of the genome-scale metabolic networks of the involved organisms was used to simulate the co-cultures and it was shown that potential metabolic interactions in co-cultures of SLAB and NSLAB may produce flavour compounds in the ripening period, possibly explaining improvements in cheese quality by regulating metabolic fluxes among microorganisms (Özcan et al., Reference Özcan, Selvi, Nikerel, Teusink, Toksoy Öner and Çakır2019; Özcan et al., Reference Özcan, Seven, Şirin, Çakır, Nikerel, Teusink and Toksoy Öner2021). Even though NSLAB can boost aroma of the cheese, directly adding adjunct may lead to off flavour formation in cheese (van Hoorde et al., Reference van Hoorde, Van Leuven, Dirinck, Heyndrickx, Coudijzer, Vandamme and Huys2010; Sahingil et al., Reference Sahingil, Hayaloglu, Simsek and Ozer2014). Hence, adjunct culture selection for cheese manufacture is important in relation to the presence of potential conflicting contributions of NSLAB, suggesting that complimentary experimental-based evaluation is necessary to determine which NSLAB could be the best. Of these experimental evaluations, VOC analysis has been the workhorse of observing aroma and taste changes by adding potential adjunct cultures in cheese manufacturing (Marilley and Casey, Reference Marilley and Casey2004; Di Cagno et al., Reference Di Cagno, De Pasquale, De Angelis, Buchin, Rizzello and Gobbetti2014; Bertuzzi et al., Reference Bertuzzi, McSweeney, Rea and Kilcawley2018b; Khattab et al., Reference Khattab, Guirguis, Tawfik and Farag2019). van Hoorde et al. (Reference van Hoorde, Van Leuven, Dirinck, Heyndrickx, Coudijzer, Vandamme and Huys2010) evaluated NSLAB Lacticaseibacillus paracasei strains for their lipolytic and proteolytic activities as well as VOC profiles in model cheese by steam-distillation extraction coupled with GC-MS. Upon adding L. paracasei strains, overall cheese flavour composition was altered by increasing aldehydes, sulphur compounds, alcohols, secondary alcohols and also by decreasing ketones and lipid-derived volatile compounds. Principal component analysis (PCA) demonstrated the obvious difference between non-adjunct culture-added cheese volatilome and adjunct culture added cheese volatilome (van Hoorde et al., Reference van Hoorde, Van Leuven, Dirinck, Heyndrickx, Coudijzer, Vandamme and Huys2010). A similar approach was undertaken to assess the aroma-producing capabilities of 11 candidate microbial strains for their potential use in cheese processing (Pogačić et al., Reference Pogačić, Maillard, Leclerc, Hervé, Chuat, Yee, Valence and Thierry2015). Volatile compounds were detected by headspace (HS)-trap GC-MS system, and the strain selection process was carried out in a curd-based medium mimicking cheese-like conditions. Overall volatilomes of cheese produced by these strains were assessed, and PCA revealed differences among strains, heralding that utilization of different microbial strains for cheese manufacturing can lead to different volatile profiles (Pogačić et al., Reference Pogačić, Maillard, Leclerc, Hervé, Chuat, Yee, Valence and Thierry2015). VOC analysis combined with sensorial analysis of cheeses made with attenuated adjunct cultures enabled the evaluation of the flavour-producing performance of different strains. There are, however, confounding results that were not correlated with VOC analysis, which showed quantitative alterations of cheese aroma compounds during the study. Due to higher amino acid content of cheese made with attenuated adjunct cultures compared to non-attenuated ones, 1-propanol, 1-hexanol, 3-methyl-2-butanol, 2-methyl-1-butanol, and 2-methyl-propanol of the control group made without adjunct culture were lower than that of the cheese made with attenuated adjunct starter (Di Cagno et al., Reference Di Cagno, De Pasquale, De Angelis, Buchin, Calasso, Fox and Gobbetti2011). Branched chain amino acids such as leucine and valine were less abundant in cheese made without adjunct culture, resulting in more VOCs produced by attenuated starter cultures than that of non-attenuated starter cultures. In this study, 10 untrained panelists evaluated cheese quality with a rubric on flavour and aroma intensity, texture and overall acceptability (Di Cagno et al., Reference Di Cagno, De Pasquale, De Angelis, Buchin, Calasso, Fox and Gobbetti2011). Without employing VOC analysis for assessing flavour-producing performances of attenuated adjunct starter cultures, the same research group used only a sensorial analysis, in which 42 panelists participated using forced-choice protocol with random distribution of cheese samples. Results showed that 45-d ripened cheese with attenuated adjunct culture reached the same sensorial level of 60-d ripened cheese that did not contain attenuated adjunct culture. Even though the activities of amino peptidase type N, cystathionine lyase were weakened due to the application of the attenuation process, the overall acceptance of cheese made with attenuated adjunct cultures was similar to the cheese made with non-attenutaed adjunct cultures (Di Cagno et al., Reference Di Cagno, De Pasquale, De Angelis and Gobbetti2012). In another study, without VOC analysis, the effects of attenuated L. lactis, Leuconostoc lactis, Vibrio casei, Staphylococcus equorum, Brochothrix thermosphacta on cheese ripening were tested via sensorial evaluation in which 11 trained panelists attended three consecutive tasting sessions where all cheese samples were randomly subjected to the sensorial analysis. Cheese made with attenuated L. lactis had the best score in terms of overall sensorial acceptance (Calasso et al., Reference Calasso, Minervini, De Filippis, Ercolini, De Angelis and Gobbetti2020). It is interesting to consider to what extent VOC analysis could corroborate/associate sensorial evaluations in these studies.
To observe VOCs-producing capabilities of NSLAB and SLAB, VOCs analyses were performed in a model medium. VOCs profiles of isolated NSLAB strains L. casei and L. rhamnosus were detected in lactose-free cheese-based medium for simulating cheese-like environment (Sgarbi et al., Reference Sgarbi, Lazzi, Tabanelli, Gatti, Neviani and Gardini2013). Also, 11 LAB strains were compared based on generating VOCs in a cheese-based medium to find and correlations between Caciocavallo Palermitano cheese volatile compounds and the LAB strains. Five LAB strains showed similar VOC profiles to the Caciocavallo Palermitano cheese VOC profile (Guarrasi et al., Reference Guarrasi, Sannino, Moschetti, Bonanno, Di Grigoli and Settanni2017), although the presence of these strains was not demonstrated in the cheese.
Stefanovic et al. (Reference Stefanovic, Kilcawley, Rea, Fitzgerald and McAuliffe2017a) reported that L. casei and L. paracasei, known as casei group members, had assocations with flavour formation during cheese ripening. The activity of enzymes involved in the proteolytic processes for flavour development (cell envelope protease, aromatic aminotransferase, and glutamate dehydrogenase) and VOC analyses in medium enriched separately with phenylalanine (as an aromatic amino acid), methionine (as a sulphur amino acid) and leucine (as a branched chain amino acid) were carried out to distinguish 10 casei group microorganisms based on their aroma-producing capability. Of all the 10 strains evaluated for flavour-forming performances in enriched medium, DPC 4206 strain was the most distinct as evident by PCA plots of VOCs for each strain in enriched medium (Stefanovic et al., Reference Stefanovic, Kilcawley, Rea, Fitzgerald and McAuliffe2017a). It is interesting to note that the DPC 4206 strain did not have distinct enzymatic activities amongst all 10 strains, indicating that enzymatic activity assays cannot help fully delineate aroma-producing potential. In parallel with that, flavour-forming abilities of Macrococccus caseolyticus subsp. caseolyticus strains were measured in a similar way. Isolated M. caseolyticus subsp. caseolyticus strains had high proteolytic and esterase activity, indicating that M. caseolyticus subsp. caseolyticus strains can contribute to flavour formation during cheese making process. Cultivated into lactose free milk, M. caseolyticus subsp. caseolyticus strains liberated more amino acids from peptides as compared to low proteolytic activity strains. Having proven high proteolytic and lipolytic activity of tested strains, VOC analysis was undertaken. As compared to the control group which was uninoculated lactose free medium, 23 VOCs out of 74 VOCs detected by headspace solid phase micro-extraction (HS SPME) were only produced by M. caseolyticus subsp. caseolyticus strains used in the study. Of 23 volatiles, methyl-hexanoate, ethyl-decanoate, 2,3-heptanedione, phenylethyl alcohol were not produced by all isolated M. caseolyticus subsp. caseolyticus strains. But, hexyl-butanoate, 2-undecanone, nonanal and benzeneacetaldehyde were produced by some isolated M. caseolyticus subsp. caseolyticus strains. For instance, benzeneacetaldehyde was produced by only two M. caseolyticus subsp. caseolyticus strains out of three M. caseolyticus subsp. caseolyticus strains. Even though isolated M. caseolyticus subsp. caseolyticus strains provided high amino acid content as well as high proteolytic and esterase activity, not allmof the distinctive volatile compounds detected in HS SPME GC-MS were the same for all M. caseolyticus subsp. caseolyticus strains (Mazhar et al., Reference Mazhar, Kilcawley, Hill and McAuliffe2020). Results of proteolytic and lipolytic enzyme activities were not sufficient to explain irregular presence of volatile compounds produced by isolated M. caseolyticus subsp. caseolyticus strains. In this study, results of enzymatic activity assays were not fully in agreement with data presented by VOC analysis.
Cheese ripening can take from two weeks up to two years. Depending on L. lactis physiological state during cheese ripening, the development of volatile compounds is endowed by metabolic pathways of aroma formation. Therefore, to analyse the aroma-producing ability, L. lactis with near-zero growth rate can be used to understand how flavour production occurs during the cheese ripening. van Mastrigt et al. (Reference van Mastrigt, Abee, Lillevang and Smid2018a) utilized retentostat cultures, in which supernatants of nutrient-poor media are continuously refreshed while keeping biomass by means of a filter in the bioreactor. This can help L. lactis reach its slow-growing physiological state, simulating that found during cheese ripening. When cultivated in chemically defined medium (CDM) containing lactose, ammonium citrate, and bacto-tryptone, slow-growing L. lactis produced 62 VOCs detected in HS GC-MS. In this study, VOC analysis was conceptually carried out in a different way. Intracellularly and extracellularly-present VOCs were identified, with the result that intracellular flavour compounds contributed to 16 VOCs produced at near-zero growth rate in CDM (van Mastrigt, et al., Reference van Mastrigt, Abee, Lillevang and Smid2018a). It can be inferred that not only would cheese microbiota contribute to the cheese flavour profile, but also cheese microbiota itself could improve cheese flavour composition through VOCs produced/remained within cells.
Another functional use of VOC analysis was performed in appraising L. lactis subsp. lactis biovar diacetylactis aroma-forming potential for 8-week slow growth in milli-model cheese. Imitated by retentostat culture, slow growth of L. lactis during cheese ripening was ensured by growing it in three different media, namely, CDM, hydrolysed micellar casein isolate and milk supplemented with 1% bacto-tryptone. In batch and retentostat cultures, 82 aroma compounds were generated. Of these 82, only 10 aroma compounds were specifically formed by retentostat cultures of L. lactis. Extensive VOC analysis of L. lactis retentostat cultures indicated that L. lactis cultivated in CDM showed an increase in some fatty acid-derived VOCs such as methyl ketones, primary and secondary alcohols, even though CDM did not contain fatty acids. When quantitively compared to milli-cheese model system, L. lactis produced more ethanol and acetaldehyde in rententostat cultures. 3-methyl-1-butanol and 2-methyl-1-butanol as branched-chain alcohols were low in retentostat cultures, suggesting that branched-chain amino acids liberated by L. lactis could be low as found in the milli-cheese model system (van Mastrigt, et al., Reference van Mastrigt, Gallegos Tejeda, Kristensen, Abee and Smid2018b).
Implementation of VOC analysis for identifying biomarkers of cheese ripening
Activity of the cheese microbiota influences the cheese metabolome during cheese ripening. Since an obvious relationship between VOCs and microbiome exists, intertwining microbiome and metabolome would explain which species and/or strains can contribute most to flavour (or even specific VOCs) during cheese ripening (Bertuzzi et al., Reference Bertuzzi, Walsh, Sheehan, Cotter, Crispie, McSweeney, Kilcawley and Rea2018a; Afshari et al., Reference Afshari, Pillidge, Dias, Osborn and Gill2020a). Effects on microbiome and VOCs production by two different mixed industrial starter cultures harbouring yeast and bacteria for surface-ripened cheese were studied for cheddar cheese ripening. Also, the intertwining of microbiome and VOCs was executed to elucidate which strains of starter cultures were associated with certain VOCs (Bertuzzi et al., Reference Bertuzzi, Walsh, Sheehan, Cotter, Crispie, McSweeney, Kilcawley and Rea2018a). Overall, 53 VOCs were detected in the cheese made with industrial starters. The number of esters and aldehydes of surface-ripened cheddar cheese were less than other VOCs detected in HS SPME GC-MS. Sulphur compounds that were not detected in the 30-d ripened cheese without starter culture were only produced in the ripened cheese made with industrial starter cultures. Also, pentanoic acid production was observed only in one starter culture in which Glutamicibacter arilaitensis was allegedly responsible for pentanoic acid production as revealed through integration of microbiome and VOC analysis (Bertuzzi et al., Reference Bertuzzi, Walsh, Sheehan, Cotter, Crispie, McSweeney, Kilcawley and Rea2018a). A similar approach was undertaken to investigate Kazak cheese microbiome and metabolome connection. In this study, microbiome and VOCs of Kazak cheese were monitored at 3-d intervals during ripening. Species-level microbiome analysis combined with VOCs analysis showed that 8 bacterial genera played significant roles to form VOCs during Kazak cheese ripening (Zheng et al., Reference Zheng, Ge, Lin, Zhang, Chen, Xiao, Wang and Shi2021). Metabolomics data obtained by untargeted GC-MS were intercorrelated with microbiome to show differences of artisanal and industrial cheese (Afshari et al., Reference Afshari, Pillidge, Read, Rochfort, Dias, Osborn and Gill2020c), and to demonstrate different cheese produced by different manufacturers with different cheese ripening periods (Afshari et al., Reference Afshari, Pillidge, Dias, Osborn and Gill2020b). Further applications of correlation analysis were performed to define biomarkers of cheese ripening. With the power of untargeted cheese metabolome data of GC-MS, cheese quality was evaluated to find biomarkers during cheese ripening. Afshari et al. (Reference Afshari, Pillidge, Dias, Osborn and Gill2021) developed a different point of view to unveil specific biomarkers of high-quality cheese. A two-step process was undertaken to find biomarkers of high-quality cheese: classifying quality of cheddar cheese by sensorial evaluations of professional cheese graders and applying data integration analysis for biomarker discovery using latent components (DIABLO). Having determined the cheese quality by professional cheese graders, microbiome and metabolome data were integrated by DIABLO. It was interesting to note that St. thermophilus was abundant in high-quality cheddar cheese while L. lactis was abundant in low-quality cheddar cheese. With respect to biomarker compounds detected in GC-MS, low-quality cheddar cheese was low in proline, histidine, aspartic acid and isoleucine and high in stearic acid and octadecanol as compared to high-quality cheddar cheese (Afshari et al., Reference Afshari, Pillidge, Dias, Osborn and Gill2021).
Malty aroma formation in raclette-type cheese is an off-flavour defect that is supposedly produced by isoleucine, valine, and leucine amino acids conversion into malty flavour compounds. Through GC-olfactometry, 3-methyl-butan-1-ol and 3-methyl-butanal were found as main indicators for formation of malty aroma in raclette-type cheese. To observe malty aroma formation, L. lactis subsp. cremoris was grown in skim milk (SM) and de Man, Rogosa and Sharpe (MRS) medium supplemented with leucine, α-ketoglutaric acid and α-ketoisocaproic acid, which are known to participate in the formation pathway of 3-methyl-butanal. Interestingly, it was shown that 3-methyl-butan-1-ol and 3-methyl-butanal production in spiked MRS and SM media were observed already during the first hours of cheese manufacture with the latter compound formed even before microbial activity when spiked with α-ketoisocaproic acid. The discrapancies between the two media pointed to the presence of other unknown reactants that might be responsible for formation of malty aroma in raclette-type cheese (Meng et al., Reference Meng, Piccand, Fuchsmann, Dubois, Baumeyer, Tena Stern and Von Ah2021).
In conclusion, aroma and taste in cheese are achieved during cheese ripening. To elucidate flavour compounds produced during cheese ripening, VOC analysis has been utilized extensively for many years. Corroborating VOC analysis results with enzymatic assays and sensorial evaluations can be useful but is cumbersome. It should also be noted that enzymatic assays in isolation maybe misleading since enzymatic activities are measured under optimum conditions for the enzyme rather than in a real cheese-like conditions. On the other hand, metabolomic data from newly applied tools such as novel liquid chromatography-based MS, nuclear magnetic resonance and untargeted GC-MS would certainly add to the understanding of the flavour formation during cheese ripening (Afshari et al., Reference Afshari, Pillidge, Read, Rochfort, Dias, Osborn and Gill2020c; Pisano et al., Reference Pisano, Rosa, Putzu, Marincola, Mossa, Viale, Fadda and Cosentino2020). Since metabolomics provides higher numbers of metabolites as compared to conventional VOC analysis, the integrated metabolome and microbiome might provide more possible biomarkers associated with specific cheese flavour traits (Fig. 2).

Fig. 2. Emerging tools to identify indicators of cheese quality and cheese ripening.
GC-MS based metabolomics approaches are becoming more common with greater sensitivity to detect a wide range of metabolites. The foremost advantage of untargeted GC-MS analysis comes from the ability to detect VOCs and non volatile compounds through derivatization (Afshari et al., Reference Afshari, Pillidge, Dias, Osborn and Gill2021).
It is our opinion that simply inputting enormous quantities of high-throughput data into understanding cheese ripening may not clear up all of the confounding results obtained by omics technology. Questions such as how to decide when cheese ripening is sensorially at its optimum and which biomarkers at what concentration indicate successful and adequate cheese ripening are still requring answers. Even though VOC analysis reflects quantities of numerous compounds for cheese ripening, there is no unanimous theory that a certain VOC profile at a certain concentration is a sign or indicator of cheese ripening for specific cheese types. Considering the wide variety of cheese types, determining indicators of the completion of cheese ripening will be a quite arduous and laborious task to achieve.




