Introduction
Noroviruses (NoVs) are single-stranded RNA viruses belonging to the family of Caliciviridae recognised as one of the most important causes of non-bacterial gastroenteritis (GE) worldwide. The genome of ~7.5 kb is organised in three open reading frames (ORF1–ORF3). Based on sequence comparisons, NoV has been classified into six genogroups (I–VI) that are subdivided into over 40 genotypes. Genogroups I, II and IV have been found to infect humans [Reference Zheng1]. The genotype GII.4 has been identified as the predominant genotype worldwide and every 2–3 years new GII.4 variants emerge [Reference Siebenga2].
The most common symptoms during NoV infection are vomiting and diarrhoea which usually resolve in 24–72 h, even if shedding of viruses may continue several days after recovery. NoVs are involved in both sporadic cases of GE and in outbreaks involving people of all age groups including children [Reference Patel3, Reference Kroneman4]. Outbreaks have been reported in different settings, especially semi-closed communities such as households, schools, hospitals, nursing homes and cruise ships. This is linked both to the increased likelihood of virus spread through person to person and to contaminated environment. Although the incidence of acute GE in adults is generally lower than in children, old age is recognised as a major risk factor for both hospitalisation and death due to GE [Reference Hutson5].
In Italy, over the last decade a group of researchers, ISGEV (Italian Study Group for Enteric Viruses, http://isgev.net), have monitored the circulation of NoV in Italian children. From January 2015 through to February 2016, 2603 faecal samples of children (0–14 years of age) with diarrhoea were investigated in three Italian areas (located in the north and south of the country) resulting in a mean NoV prevalence of 12.1% (316/2603) [Reference Giammanco6]. Besides these data, epidemic outbreaks of GE are largely under-reported, data on the adult population are lacking and diagnosis of NoV is performed rarely in Italy. As a consequence, only few outbreaks related to NoV have been described to date [Reference Di Bartolo7–Reference Rizzo10]. Although the same genotypes circulating in humans in Europe are frequently detected in food (mussels and ready to eat vegetables) in Italy [Reference La Rosa11–Reference La Bella13], in the absence of an accurate surveillance, the role of this virus in the aetiology of GE is unnoticed by local public health offices.
In this paper, we report the results of a pilot surveillance programme that was run for 2 years (January 2012 to December 2013) with the specific aim of implementing the active surveillance of sporadic cases and epidemics in communities caused by NoV in the autonomous province of Bolzano in Italy.
Methods
Sample collection
This study was conducted in the Unit of Infectious Diseases Molecular Biology (Laboratory for Microbiology and Virology, Bolzano Hospital) of autonomous province of Bolzano (7400 km2; 515 714 residents). The Unit is the reference centre for microbiology and virology analyses for the whole area. Every year, over 13 000 stool samples are submitted for microbiological analyses from the local sanitary authority, general practitioners, short-term care facilities and from the seven hospitals of the same area (total: 1843-bed hospitals; 64 000 discharges). The 702 patients enrolled in this study were affected by GE (defined as three or more loose stools within a 24 h period), with or without vomiting. In the case that multiple stool samples were collected from the same patients, those submitted within a 12-day period were considered part of the same episode and considered as one case. Patients were considered as outpatients if stool samples were obtained from subjects attending the Accident & Emergency department (A&E unit) or visiting their general practitioners. Cases were defined as nosocomial if patients were not admitted at the hospital for GE and symptoms occurred >72 h after the admission. In this study, cases from outbreaks were also included. If there were at least three cases with GE linked by time and location, the episode was considered to be an outbreak.
Information on symptoms, age and residence was collected with a specific questionnaire administered to volunteer patients affected by GE and positive for NoV. Since questionnaires were not routinely administered, limited information was available. All samples were collected following informed consent of the patients and for children from parents or guardians.
Faecal samples were stored at −20 °C until examination.
Detection and genotyping of NoV
Total DNA/RNA was extracted from 10% faecal suspension (w/v) using the NucliSens easyMAG instrument and reagents (bioMerieux, France), following manufacturer's instructions. NoV RNA was detected by real-time reverse transcription PCR (RT-PCR) assay that enables to differentiate between GI and GII NoVs with genogroup-specific primer sets [Reference Kageyama14]. RNA from samples positive for NoV by real-time RT-PCR was tested by RT-PCR, using two sets of primers, G1SKF/G1SKR or G2SKF/G2SKR specific for either GI or GII, that anneal to ORF2. Finally, a selected group of samples was further tested by one-step RT-PCR using primers JV12 and G2SKR or G1SKR, which amplify an 1100nt fragment overlapping the ORF1–ORF2 regions of genogroup II NoV [Reference Vinje and Koopmans15, Reference Kojima16]. This step was followed by a semi-nested PCR, using the JV12 and JV13 primers that anneal in the ORF1 [Reference Vinje and Koopmans15].
The DNA amplicon of expected size were sequenced by BioFab S.r.l. (Italy). Sequences obtained were analysed and edited using the Bionumerics software packages v6.0 (Applied Maths, Belgium).
The sequences obtained were aligned with those present in the NCBI GenBank (http://www.ncbi.nlm.nih.gov/), genotypes and variants were assigned using the public database Noronet typing tool (http://www.rivm.nl/mpf/norovirus/typingtool). Phylogenetic analyses were carried out using neighbour-joining with the parameters suggested by the model test, using the Molecular Evolutionary Genetic Analysis (MEGA 6) software [Reference Tamura17]. Confidence values at the nodes were obtained by performing 1000 bootstrap analyses.
The GenBank accession numbers for sequences obtained are: KF475926–KF475961; KF738671–KF738694 JC; KF475962–68; KF475924–25.
Statistical analyses
Patients were arbitrarily divided into four categories based on the quartile distribution of age (⩽20, 21–43, 44–68, ⩾69 years old). Qualitative data were analysed using χ 2 test. All statistical analyses were performed using the software SPSS 23 (SPSS Inc., Armonk, NY, USA).
Results
During the 24-month period (January 2012 to December 2013), 702 stool samples were submitted to the reference centre for microbiology and virology (autonomous province of Bolzano, Italy) from patients (353 female; 349 male; median age: 44 years) affected by GE.
Of the 702 patients, 287 (2012: 82/254; 2013: 205/448) were seen as outpatients (either clinics or A&E departments) and 415 (2012: 172/254; 2013: 243/448) were community cases requiring hospitalisation or hospital-acquired GE.
All 702 samples met the inclusion criteria and were submitted to testing for NoV diagnosis. Of the 702 samples, from unique patients, 162 were positive for NoVs by real-time RT-PCR. Twenty-five stool samples from the patients were followed up, collecting one additional sample after a mean of 13 days from people who were positive for NoV at the first test and 12 out of 25 were again confirmed as positive.
Among the 162 NoV-positive cases, 37 were hospital-acquired, 49 were community-acquired sporadic cases and 76 GE cases occurred in the same settings and in the same days were grouped in nine suspected outbreaks.
The monthly distribution of GE cases and the detection rate of NoVs are shown in Figure 1.
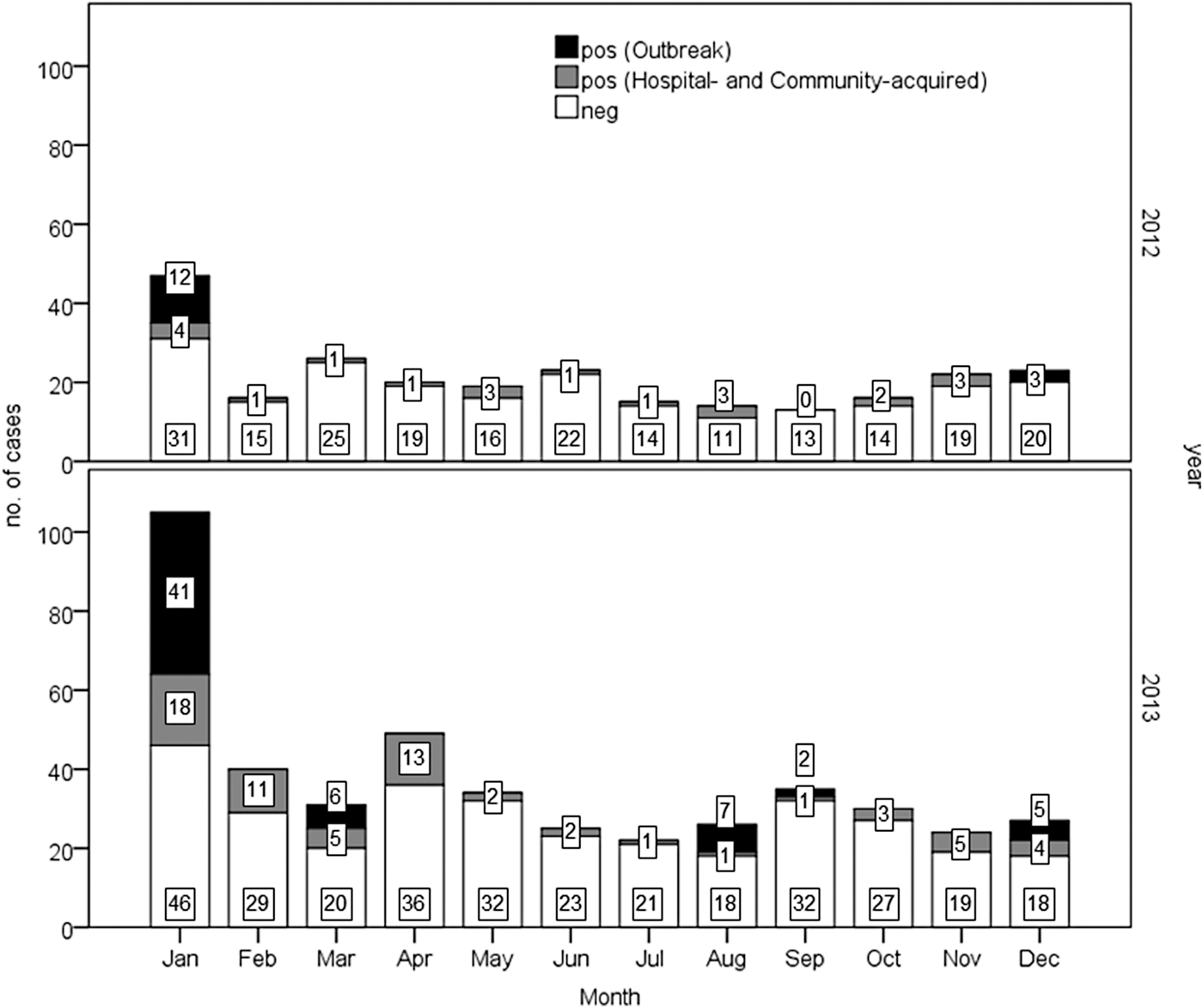
Fig. 1. Monthly distribution of acute gastroenteritis cases in 2012–2013. Results of norovirus detection among patients are reported (positive in black and grey).
Overall, a significantly (P < 0.001) higher prevalence of NoV infections was observed in 2013 (28.3%; 127/448) than 2012 (13.8%; 35/254). Similarly a higher prevalence (P < 0.01) was detected in 2013 (20.6%; 66/321 vs. 9.1%; 20/219) considering only hospital- and community-acquired cases.
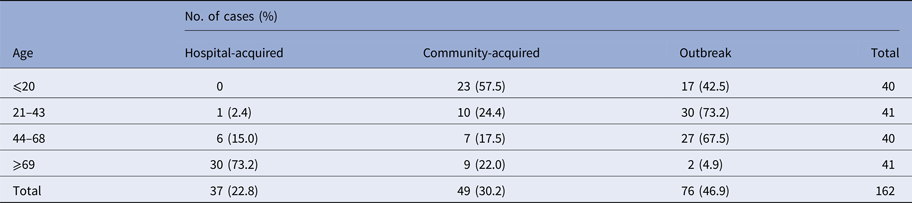
A similar annual trend was observed in the occurrence of NoV-positive sporadic cases in both community- (49 cases) and hospital-acquired (37) cases, with a peak in January in both years (data not shown). NoV infections were found in all age classes in outbreak and community-acquired cases, while hospital-acquired infections occurred in people older than 44 years (one case in a subject 37 years old), with the highest peak in elderly (⩾69 years; 30/37) (Table 1). Eighteen cases in subject younger than 5 years were positive for NoV. In the studied period, 80 cases of GE were reported in this age group, 21 of which (26%) were positive for rotavirus (data not shown). The average annual incidence rates/100 000 residents (131 cases) (95% CI) are the following: 3.67 (3.16–4.24) for hospital-acquired, 4.06 (3.53–4.66) for nosocomial, 7.73 (6.98–8.53) for hospital- and community-acquired.
Table 1. Number of cases and prevalence of norovirus correlated to both age classes and epidemiology of infection
Based on real-time RT-PCR results, which enable to distinguish between GI and GII, the most common genogroup was GII, accounting for 127 out of 162 (78.4%) cases tested. Strains belonging to GII were identified in 49 community-acquired sporadic cases (GI was detected in one sporadic case) and in all the nosocomial cases. In six out of nine outbreaks studied GII NoVs were identified.
Genotyping identification was based on partial sequencing of the ORF2 gene (capsid). Because of budget limitations, it was conducted on 43 sporadic cases and 29 cases linked to the outbreaks investigated.
Analyses of the resulting sequences of sporadic cases revealed six different capsid genotypes involved. The most prevalent genotype was GII.4 (36/43; 83.7%), followed by GII.3, GII.1 and GII.6 (5%), which were detected in one or two cases per year, and GII.2 detected in only one case (Table 2). Genotyping showed that NoV GII.4 was the most commonly identified type, accounting for all nosocomial cases (Table 2) (21 sequences obtained). Sequence alignments indicate that GII.4 strains belonged to the two variants GII.4 Sidney 2012 and GII.4 New Orleans 2009. Sequence comparisons revealed a close relatedness (≈99%) with human NoV strains (e.g. KF668569, KP244322) detected in the paediatric population in the same years in Italy.
Table 2. Genotype distribution of norovirus among CA and NOS cases in 2012–2013
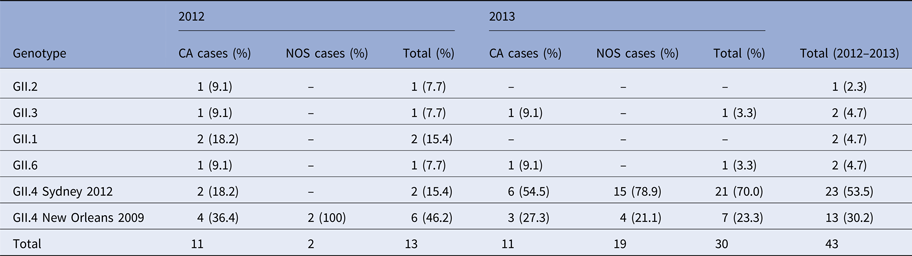
CA, community acquired; NOS, nosocomial cases.
During 2 weeks in January–February 2013, four cases in the elderly ward were linked to the same (100% n.i.) GII.4 Sidney 2012 strain (Acc. No. KF738680-82-83-88) (Fig. 2). Similarly during 2 weeks in March–April 2013, another GII.4 Sidney 2012 (Acc. No. KF738687-89; 100% n.i.) strain displaying a 100% nucleotide identity with the earlier strain, affected three patients of the general ward. During the same period in both wards, a total of 22 cases of GE were detected and linked to GII NoV. These data suggest that small clusters might have occurred in these wards, since the same viruses sharing 100% nucleotide identity were detected in patients in the same wards (Fig. 2).
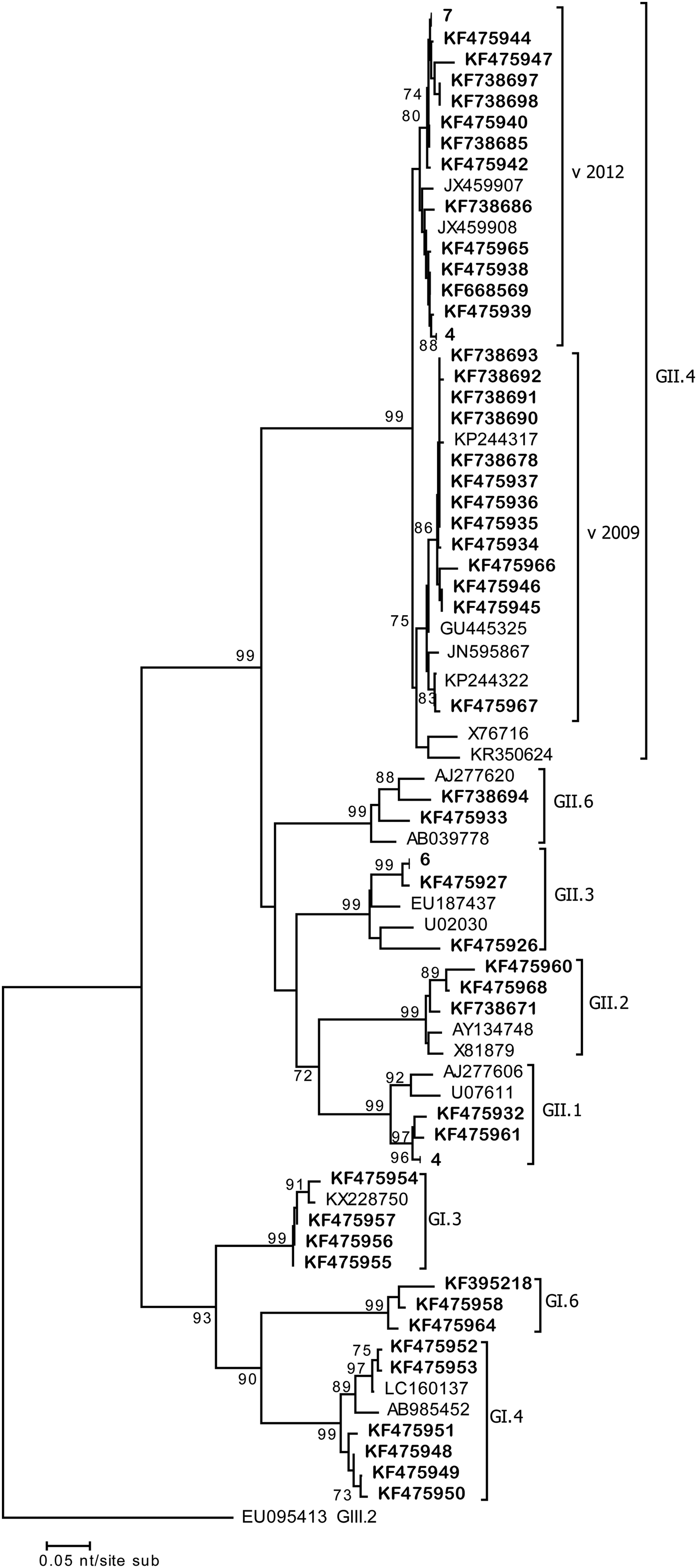
Fig. 2. Phylogenetic tree (NJ) based on the 290 bp sequences of the ORF2 fragment. Representative strain sequences of different genotypes are included. Strains detected in this study are in bold. Those sequences displaying 100% identity were grouped together; number of sequences grouped is indicated. Bootstrap values >70 are indicated.
Among community-acquired cases, those which occurred in the same setting and at the same time were grouped as suspected outbreaks. No epidemiological study was conducted, so no information is available on the suspected outbreaks. A total of nine outbreaks were identified, with a peak between December 2012 and March 2013 (six outbreaks). Four of them occurred in restaurants, two in canteens, one in a kindergarten, one in a long-term care institution and one in a secondary school. However, no information was available on the route of transmission or the source of infection, due to a lack of epidemiological data.
Overall, 76 cases involved in the outbreaks, tested positive for NoV by real-time, 33 and 43 genogroup I and II, respectively. GII was the most prevalent accounting for six out of the nine outbreaks studied. Twenty-five samples, at least two samples for each outbreak, were genotyped by sequencing the capsid partial gene (Table 3). For three outbreaks, no sequences are available, and only the genogroup was identified by real-time RT-PCR.
Table 3. Genotypes distribution among NoV outbreaks
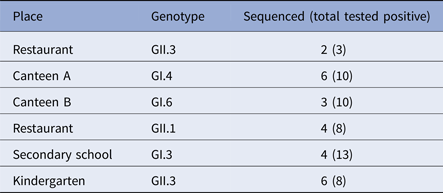
No GII.4 strains were revealed among outbreak samples sequenced. GII.3 NoV was involved in outbreaks occurring in restaurants and the kindergarten, GI.4 and GI.6 in two outbreaks occurred in canteens, and GII.1 in a restaurant. Sequence alignment showed that the same strain (100% nucleotide identity) was involved in each of the outbreaks investigated, suggesting that a common source of infection was possible.
Since recombination is common with NoV, we investigated a subset of samples for evidence of a possible inter-genotype recombination by sequencing the region spanning the ORF1/ORF2 or by separately sequencing both the partial capsid gene (ORF2) and RdRp (ORF1).
The sequence analyses revealed several recombinant strains, i.e. GII.P7_GII.6, GII.Pg_GII.1, GII.P16_GII.3, gIPb_GI.6. A subset of GII.4 variants was also analysed; three strains of Sydney variant 2012 possessed the original gII.Pe, two possessed the GII.P4 ORF1 that clustered with the New Orleans 2009. The remaining eight NoV strains were identical in both ORF1 and ORF2 fragments and were: GII.P2_GII.2, GII.P3_GII3, GII.P6_GII.6, GI.P4_GI.4, GII.P4 New Orleans 2009_GII.4 New Orleans 2009.
Discussion
To date, only few outbreaks of GE associated to NoV have been described in detail in Italy [Reference Giammanco9, Reference Rizzo10, Reference Di Bartolo18, Reference Le Guyader19]. More recently, several studies on sporadic NoV cases in the paediatric population have been reported [Reference Giammanco6, Reference Colomba20, Reference Ramirez21], showing that as described worldwide, this enteric pathogen is a common cause of acute GE requiring hospitalisation among children but also an important cause of nosocomial cases that can determine the occurrence of hospital outbreaks [Reference Valentini22].
This study presents data on a 2-year surveillance of patients affected by GE, both adults and children, in urban and rural areas of the autonomous province of Bolzano in northern Italy. In this study, the average annual incidence rate/100 000 residents of NoV infections in sporadic cases was 7.73 and 23% of all cases of GE investigated were caused by NoV. The incidence of sporadic cases is lower than reported in other countries [Reference O'Brien23], which generally refer to a wider population. This study has a sampling bias caused by inclusion of samples collected for routine diagnostic virus analyses, and GE cases are frequently under-reported, particularly those which are community-acquired. Therefore, the studied population does not encompass all cases of acute GE, which is the likely explanation of the low incidence observed.
Our study also confirms that hospitals represent an important setting for NoV infections, which are more frequently associated with GII.4 genotypes in older people, as reported in several studies in Europe [Reference Kambhampati24, Reference Franck25].
The ability of NoV to spread efficiently within hospitals among the elderly, who were the nosocomial patients affected by GE in this study, is likely due to the physical weakness of patients, their prolonged length of stay in the hospital and the exposure to incoming infectious agents via guests or visitors. In this study, long symptomatic periods of up to 30 days, with a mean duration of 18 days, were observed in the follow-up of patients. Recurrent infections during the same year were also detected in two immunosuppressed patients (data not shown). Detection of individuals shedding NoV for extended periods of time confirms that patients may act as reservoirs for the spread of the virus to other susceptible people, especially in confined areas.
Circulation of different genotypes was observed. The most common was GII.4 with the GII.4 New Orleans 2009 and Sydney 2012 being the only two variants detected, as previously seen in Italy in the same years in the paediatric population [Reference Giammanco26]. Conversely GI was only detected in two outbreaks.
In conclusion, our present study shows that an implemented surveillance of GE in communities and in hospitals, together with enhanced diagnostic protocols, may detect a relevant number of both sporadic cases and outbreaks associated to NoV infections. NoV probably circulates widely in Italy, as in other European countries but its incidence is largely underestimated. Data obtained in this study were used to implement guidelines for NoV management (e.g. diagnostics, hand hygiene, patient transfer and training of health care staff) in the autonomous province of Bolzano, determining a reduced number of nosocomial cases in the subsequent years. Active surveillance is still ongoing.
Acknowledgements
This study was partially supported by grants from the Ministry of Health, Italy: ‘Creazione di una rete di laboratori virologici di riferimento (Noronet-talia) per il controllo delle epidemie di gastroenterite da Norovirus in Italia’, CCM 2009. The authors kindly thank Dr Silvia Spertini and Dr Martina Born for their contribution to collecting data.
Declaration of interest
None.






