DNA methylation occurs when a methyl group (CH3) is added to the carbon-5 position of cytosine in CpG dinucleotides(Reference Jin, Li and Robertson1). Methylation of gene promoters and regulatory regions hinders the binding of transcription factors – this is the mechanism of epigenetic gene regulation(Reference Pauwels, Ghosh and Duca2). The global DNA hypomethylation and region-specific hypermethylation are known as one of the epigenetic hallmarks of cancer(Reference Deng, Nguyen and Tanaka3). This view is strongly supported by the finding that DNA methylation induces silencing of tumour suppressor genes(Reference Merlo, Herman and Mao4). There is a growing research interest in determining whether changes in the global status of DNA methylation are related to the environment and whether these changes can be biomarkers of disease(Reference Nelson, Marsit and Kelsey5). The researchers have not yet reached a consensus on the meaning of global DNA methylation. However, a reasonable definition is the measure of methylcytosine content of the genome overall, of repetitive elements or of multiple gene regions, not considering specific genes or regulatory regions(Reference Nelson, Marsit and Kelsey5).
Increasing evidence indicates that DNA methylation is labile in response to nutritional status(Reference Brunaud, Alberto and Ayav6). Nutrients naturally provided in the diet such as methionine, folate, betaine, choline and vitamins B2, B6 and B12 are methyl donor precursors in one-carbon metabolism(Reference Anderson, Sant and Dolinoy7), and genetic variants, such as polymorphisms in methylenetetrahydrofolate reductase (MTHFR), methionine synthase (MTR), MTR reductase and other enzymes, can alter this metabolism(Reference DeVos, Chanson and Liu8). One-carbon metabolism plays a central role in DNA methylation, since it determines the flux of methyl groups toward methylation of DNA. These nutrients participate in the methionine cycle for the synthesis of S-adenosyl methionine, which is the universal methyl donor for DNA methylation reactions. When S-adenosyl methionine donates its methyl group, it is converted to S-adenosyl homocysteine(Reference Anderson, Sant and Dolinoy7). In parallel, an important nutrient-dependent reaction is the methylation of deoxyuridylate to thymidylate in the formation of DNA by folate which is required for proper cell division(Reference Crider, Yang and Berry9).
Folate is an essential water-soluble B vitamin, also known as vitamin B9. Folate is the form of the vitamin found naturally in foods, whilst folic acid (FA) is the synthetic form available in supplements and used for food fortification. Currently, eighty-six countries worldwide – including Brazil since 2002 – have implemented mandatory FA fortification of at least one cereal grain for the prevention of neural tube defects(10). The role of folate in cancer progression has been researched for a long time; however, there is no consensus on the relationship between folate and carcinogenesis, although substantial efforts have been made to determine the cause and role of this relationship(Reference Giovannucci11–Reference Kim, Smith-Warner and Spiegelman13). In this regard, epigenetic modifications, particularly DNA methylation in selected gene promoters, are recognised as common molecular alterations in human tumours and the association between folate and DNA methylation has also been studied(Reference Crider, Yang and Berry9, Reference Kim, Lee and Sidransky14).
In the present study, we aimed to investigate the effect of dietary methyl-group donor intake (methionine, folate, choline, betaine, vitamins B2, B6 and B12), biomarkers (total folate, unmetabolised FA (UMFA), 5-methyltetrahydrofolate (5-MTHF), homocysteine (hcy), vitamins B6 and B12 concentrations) and genetic variants (polymorphisms involved in one-carbon metabolism) on global DNA methylation in a population exposed to mandatory flour fortification with FA.
Materials and methods
Study population
Participants in the present study were drawn from an earlier study, namely, ‘Healthy Survey of São Paulo’ (ISA-Capital)(Reference de Carvalho, César and Fisberg15), a cross-sectional study of health and living conditions among a representative sample of residents in São Paulo, south-eastern Brazil in 2008. The survey used a complex, stratified, multistage probability cluster sampling design. This complex probabilistic sample was obtained by conglomerates in two stages: census tracts and household sectors, using data from the ‘National Household Sample Survey’ conducted in 2005(16). Six sample domains were defined by age group and sex: females and males aged 13–19 years old (adolescents), 20–59 years old (adults) and 60 years old or over (older adults). A sample size of 300 in each domain was estimated to be the minimum, based on a prevalence of 0·5 with a standard error of 0·07 at a 5 % significance level and a design effect of 1·5. The recruitment process was described in more detail in a previous study(Reference de Carvalho, César and Fisberg15).
A total of 2691 individuals, aged 12 years or over, were selected to answer questions about dietary intake, lifestyle factors (e.g. physical activity, smoking and use of drugs) and socio-demographic characteristics (e.g. age, sex, self-reported race, educational level and family income). Among these participants, 750 individuals accepted and provided blood samples, completed the dietary measurement and collected anthropometric measures. The information on measures of global DNA methylation was not available for all participants, generating a final sample size of 442 individuals.
The study protocol was reviewed and approved by the Ethics Committee at the School of Public Health, University of São Paulo (approval number: 2001). A written informed consent form was obtained from all participants.
Questionnaire data
Household information was obtained using structured questionnaires applied by previously trained interviewers. Demographic factors (e.g. age, sex and self-reported race), socioeconomic (e.g. household per capita income, educational attainment), lifestyle (e.g. physical activity level, smoking status, alcohol consumption), self-reported morbidity, family history diseases, dietary supplements, use of drugs and food intake variables were collected. The self-reported race was defined as the ethnicity characteristics self-declared by the Brazilian population according to the following options: white, black, asian, mixed or indigenous(17). After that, it was categorised as white and non-white due to the socio-economic inequalities observed among these groups.
In a subsequent home visit, blood samples were collected and anthropometric measurements including weight, height and waist circumference were performed by one nurse technician. Afterwards, BMI was calculated by dividing the measured weight by the height squared.
Dietary intake
Two 24-h dietary recalls were performed as a dietary survey. The first was collected face-to-face by interviewing the relevant households, and the second was collected telephonically. These 24-h dietary recalls were collected using the multiple-pass method(Reference Guenther, DeMaio and Ingwersen18) and automated multiple-pass method(Reference Blanton, Moshfegh and Baer19), respectively, on non-consecutive days, regardless of day of the week and season. The multiple-pass method makes use of a standardised process structured in five steps (quick listing, forgotten list foods, naming meals, detail cycle and general review) that keeps individuals interested and engaged in the interview, which helps them remember all items consumed.
Food consumption described in both recalls was converted into energy and nutrient values using the Nutrition Data System for Research (NDSR®, version 2007, Nutrition Coordinating Center, University of Minnesota, Minneapolis, MN, USA), which is based on data from the United States Department of Agriculture Food Composition Table. The nutritional adequacy of the food consumption data was verified using a national food composition table – ‘Tabela Brasileira de Composição de Alimentos’(20). Moreover, the Multiple Source Method, that is, a statistical modelling technique, was used to measure folate intake in order to estimate the usual consumption distribution of nutrients, mitigating the effects of intra-individual variation when at least two dietary measurements per individual are available.
Sample collection and biomarker analyses
For molecular and biochemical analyses, whole blood was obtained by venepuncture after a 12 h overnight fast by a trained nurse technician. Approximately 20 ml of blood was collected, in tubes containing EDTA and plastic serum tubes which had spray-coated silica. The tubes were stored in Styrofoam packages with recyclable ice packs and were transported to the Laboratory of Human Nutrition at School of Public Health, followed by centrifugation and processed into aliquots of serum and plasma, and stored in a freezer at −80°C until analysis. Total folate, that is, circulating FA plus 5-MTHF was determined in plasma samples at the Vitamin Metabolism Laboratory/Jean Mayer United States Department of Agriculture HNRCA/Tufts University by a modified affinity-HPLC method with electrochemical detection in order to measure the different vitamin forms in the blood(Reference Kalmbach, Paul and Selhub21). Vitamin B6 levels were analysed by HPLC with fluorometric detection using the ImmunDiagnostik AG® HPLC-Analytik system(Reference Rybak, Jain and Pfeiffer22). Vitamin B12 concentrations were determined by a chemiluminescence immunoassay with paramagnetic particles for the quantitative determination of vitamin levels in human serum and plasma using the Access Immunoassay System® (Beckman Coulter, Inc.)(23). The immunoassay method of chemiluminescence microparticles using the ARCHITECT Homocysteine Reagent Kit (Abbott Diagnostics Division) was used to analyse the plasma concentrations of hcy(24). All tests were performed according to the manufacturer’s instructions.
DNA extraction and genetic analyses
DNA was extracted and isolated utilising a salting out method(Reference Miller, Dykes and Polesky25). Subsequently, the DNA concentration was determined using the NanoDrop® 1000 Spectrophotometer. The polymorphisms were genotyped by allele-specific PCR amplification using the TaqMan Open Array (Life Technologies Corporation)(Reference Myakishev, Khripin and Hu26). We analysed C677T (rs1801133) and A1298C (rs1801131) for MTHFR gene, A2756G (rs1805087) for MTR gene, A66G (rs1801394) of MTR reductase – MTRR gene, G80A (rs1051266) for reduced folate carrier 1 – RFC1 gene and 19-bp deletion polymorphism for dihydrofolate reductase – DHFR gene.
In the present study, the genotyping call rate was >97 % for all the polymorphisms. For each polymorphism in the population, the minimum allele frequency was calculated and the Hardy–Weinberg equilibrium was verified.
Global DNA methylation measurements
Global DNA methylation was estimated using hydrolysis of genomic DNA followed by specific detection and quantification of the 5-methylcytosine content. First, DNA (5 µg) containing 0·1 mm deferoxamine was hydrolysed with Tris buffer including 200 mm HCl/MgCl2 (pH 7·4) and DNAse 1 (Sigma Aldrich, EUA) for 1 h at 1000 rpm and 37°C. DNA was incubated with phosphodiesterase (0·0005 units) (Sigma Aldrich) and alkaline phosphatase (1·2 units) for 1 h at 37°C. After that, the final volume (60 μl) was centrifuged for 10 min at 3000 rpm and the supernatant was collected. Aliquots of hydrolysed DNA were injected into the HPLC with diode-array detection analytical system (Shimadzu Corporation). Last, the percentage of the global DNA methylation was estimated. Other additional information about the method was described in a different study(Reference de Oliveira Andrade, Fontelles and Rosim27).
Statistical analyses
First, data were tested for normal distribution using a skewness–kurtosis test. Continuous variables were expressed as means, standard deviations, percentiles and 95 % CI. Categorical variables were expressed as numbers of subjects and percentages. Second, an independent t test and ANOVA were performed to assess the difference in global DNA methylation means according to the demographics and polymorphism characteristics. Third, Pearson’s correlations were calculated for nutrient intakes and biochemical measurements and global DNA methylation.
We assessed changes in the percentage of global DNA methylation using a multiple regression model with methyl-group donor intake as an explanatory variable. Potential confounders were selected based on their association with global DNA methylation and population nutrition: age (12–19, 20–59 and 60 or more years), sex (male and female), smoking status (non-smoker or former and current smoker), self-reported race (white and non-white), dietary intake (specific for each final model) and hcy levels (µmol/l).
All tests were two-sided, and a 5 % significance level was assumed. Statistical analyses have been performed using STATA® software (version 13.0, 2013).
Results
Of the 442 participants in the present study, the majority were women (56·8 %); in the age range 20–59 years (48·6 %); self-reported white race (60 %); non-smokers (82·1 %) and either overweight or obese (49·3 %). A significant difference in mean global DNA methylation was only observed by age group. The mean of global DNA methylation was lower in young people than in adults and the elderly (P = 0·049). The frequencies of mutant homozygotes and heterozygotes were 53·9 % for MTHFR 677C>T, 41·6 % for MTHFR 1298A>C, 34 % MTR 2756A>G, 70·2 % for RFC1 80G>A, 65·9 % for MTR reductase 66A>G and 71·1 % for dihydrofolate reductase deletion. No differences between genotypes of polymorphism and global DNA methylation mean were identified. Other information of demographic and genetic variant characteristics in the study population is shown in Table 1.
Table 1. Characteristics of subjects and global DNA methylation according to the characteristics
(Numbers of subjects and percentages; mean values and 95 % confidence intervals)
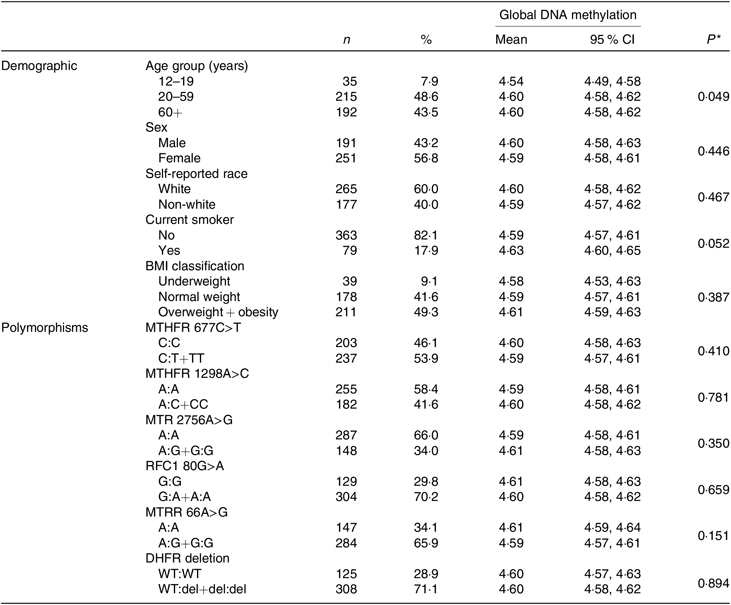
MTHFR, methylenetetrahydrofolate reductase; MTR, methionine synthase; RFC1, reduced folate carrier 1; MTRR, MTR reductase; DHFR, dihydrofolate reductase; WT, wild type; del, deletion.
* P value for t test or ANOVA.
In Table 2, we described the means and percentiles of nutrient intakes including folate (food folate, FA and dietary folate equivalents), vitamins B2, B6 and B12, choline, betaine and methionine, as well as, biochemical measurements of total folate, 5-MTHF, circulating FA, vitamins B6 and B12, hcy, global DNA methylation (%). The mean of folate intake was 361·83 (sd 107·19) and range of 284·43–423·22 (percentile 25–percentile 75) µg of dietary folate equivalents/d. Detectable amounts of circulating FA were identified in the population. The mean was 2·6 (sd 4·38) and range of 0·62–2·96 (percentile 25–percentile 75) nmol/l. The mean of global DNA methylation in percentage was 4·60 (sd 0·15) and range of 4·52–4·69 (percentile 25–percentile 75).
Table 2. Nutrient intake and biochemical measurements
(Mean values, standard deviations and percentiles)
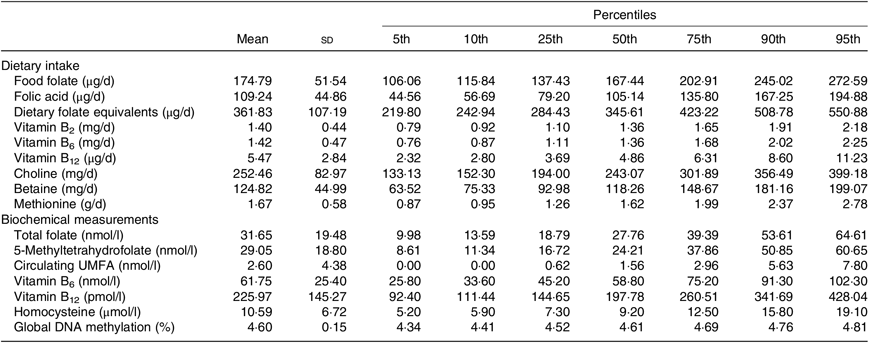
UMFA, unmetabolised folic acid.
The correlations between global DNA methylation, intake of dietary methyl-group donors and biochemical status are presented in Table 3. A significant correlation was found only for betaine intake (P = 0·040). No associations were found between biochemical markers and global DNA methylation. When we analysed the association between global DNA methylation and intake of dietary methyl-group donors using multiple regression, we observed that the increase in betaine intake led to an absolute change in percentage of DNA methylation (β = 0·0005, P = 0·024). Intake of the other nutrients did not have a significant effect on global DNA methylation. The parameters for each final model are presented in Table 4.
Table 3. Pearson’s correlations between global DNA methylation, dietary intake and biochemical measurements
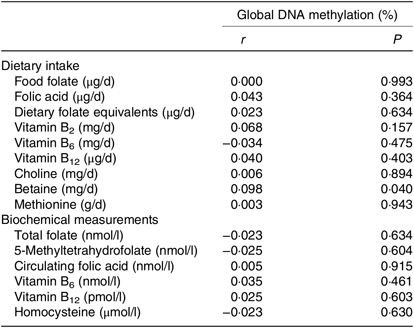
Table 4. Associations between global DNA methylation and methyl-group donor intake
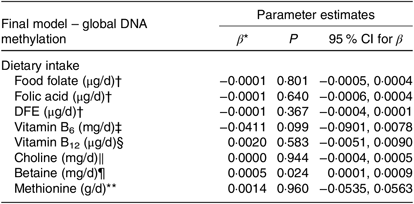
DFE, dietary folate equivalents.
* β-Estimate is an absolute change in percentage of DNA methylation.
† Model is adjusted for age, sex, smoking, self-reported race, dietary intake (betaine, methionine, choline, vitamins B6 and B12 and energy) and homocysteine.
‡ Model is adjusted for age, sex, smoking, self-reported race, dietary intake (DFE, methionine, choline, betaine, vitamin B12 and energy) and homocysteine.
§ Model is adjusted for age, sex, smoking, self-reported race, dietary intake (DFE, methionine, choline, vitamin B6 and betaine and energy) and homocysteine.
| Model is adjusted for age, sex, smoking, self-reported race, dietary intake (DFE, methionine, betaine, vitamins B6 and B12 and energy) and homocysteine.
¶ Model is adjusted for age, sex, smoking, self-reported race, dietary intake (DFE, methionine, choline, vitamins B6 and B12 and energy) and homocysteine.
** Model is adjusted for age, sex, smoking, self-reported race, dietary intake (DFE, betaine, choline, vitamins B6 and B12 and energy) and homocysteine.
Discussion
The findings of our study conducted in a population exposed to mandatory FA fortification demonstrated that global DNA methylation varied only by age and were not significantly affected by other variables, such as sex, self-reported race, smoking, BMI and polymorphisms of one-carbon metabolism genes. Global DNA methylation and dietary betaine intake were significantly and positively correlated, which was corroborated by the association between the increase in betaine intake and absolute change in percentage of DNA methylation.
DNA methylation is one of the epigenetic events extensively studied within one-carbon metabolism. Nutrients obtained through the usual diet have a direct effect on this metabolism and consequently on DNA methylation, since they influence the supply of methyl groups and the biochemistry of methylation processes(Reference Davis and Uthus28). Methionine is continuously regenerated from hcy. This essential amino acid is a substrate for S-adenosyl methionine, the principal methyl donor for DNA methylation. Betaine can be obtained through both dietary intake and the oxidation of its precursor choline, acting as a substrate in the betaine–hcy methyltransferase reaction(Reference Crider, Yang and Berry9, 24). However, folate has a crucial role in the complex one-carbon metabolism, which involves several enzymes, metabolites, substrates and cofactors. After absorption in the intestine and/or liver, folate is metabolised to 5-MTHF (Reference Crider, Yang and Berry9). This form of folate will donate its methyl group to the re-methylation of hcy to methionine, which also requires vitamin B12(Reference Anderson, Sant and Dolinoy29). But, FA in fortified foods and supplements must first be reduced to dihydrofolate and then to tetrahydrofolate by the enzyme dihydrofolate reductase in order to be incorporated into one-carbon metabolism(Reference Field and Stover30). An excessive intake of FA may exceed the capacity of the enzyme, which was also reported to be slow and easily saturated in humans, leading to the appearance of a non-metabolised fraction of FA, commonly named UMFA or circulating FA(Reference Bailey and Ayling31). Detectable concentrations of UMFA have been observed in countries after mandatory or voluntary fortification with FA, including Brazil(Reference Steluti, Reginaldo and Selhub32).
Although our study did not find associations between folate intake (such as dietary folate equivalents, FA and food folate), folate biomarkers (such as total folate, UMFA, 5-MTHF) and global DNA methylation, it is well-established that inadequate folate availability has a direct impact on DNA methylation. Low folate status has been associated with hypomethylation and genomic uracil misincorporation(Reference Linhart, Troen and Bell33). On the other hand, high FA intake, from the consumption of fortified foods and supplements, can lead to changes in DNA methylation. A study in normal human cells in vitro reported that supra-physiological concentrations of FA perturbed the intracellular S-adenosyl methionine:S-adenosyl homocysteine ratio and induced aberrant DNA methylation, with increased gene-specific CpG island methylation and decreased LINE-1 methylation(Reference Charles, Johnson and Belshaw34). In addition, the discussion of food fortification with FA, global DNA methylation, is influenced by an interaction between fortification period and erythrocyte folate concentrations among postmenopausal women. In the pre-fortification period, women with higher (v. lower) erythrocyte folate concentrations presented higher mean global DNA methylation (5·12 v. 4·99 %; P = 0·05); however, lower DNA methylation in the post-fortification period (4·95 v. 5·16 %; P = 0·03)(Reference Bae, Ulrich and Bailey35). We hypothesised that we would not find an association between DNA methylation, folate and its different forms because the whole population was exposed to mandatory fortification with FA. So, we observed homogeneous distribution of FA intake among the population if we considered that most of the population showed the presence of UMFA (>80 %). The presence of UMFA may indicate adequate folate status and possible excessive intake of FA because UMFA is only identified with saturation of the folate metabolism(Reference Obeid and Herrmann36). However, although a detectable level of UMFA has been observed in participants exposed to fortification, folate concentrations are not as high as those of supplement users(Reference Pfeiffer, Sternberg and Fazili37).
Besides folate, other nutrients known as methyl donor precursors (such as vitamins B2, B6 and B12, choline, betaine and methionine) are related to one-carbon metabolism and have also been implicated in the regulation of DNA methylation and DNA synthesis(Reference Anderson, Sant and Dolinoy29). A cross-sectional study with more than 5000 adult participants assessed the nutrient intake from a FFQ and found an association with blood DNA methylation. Low riboflavin intake was associated with higher methylation at CpG cg21230392 in the first exon of Prominin 1, that is, CpGs in gene promoters. Medians (interquartile range) for betaine intake were 356 (247·5) mg/d and 368 (249·5) mg/d for controls and cases, respectively(Reference Chamberlain, Dugué and Bassett38). Another study showed that long-term supplementation with FA and vitamin B12 in elderly subjects resulted in effects on DNA methylation of several genes(Reference Kok, Dhonukshe-Rutten and Lute39). In addition a large cohort study prospectively evaluated nutrient intake using a FFQ and self-report of supplement use(Reference Dhana, Yen and Li40). A higher intake of folate (from food only, not total folate) was associated with increased melanoma risk, while this association was not sustained when considering higher intake of vitamins B6 and B12, choline, betaine and methionine. In our study, the increase in betaine intake was associated with an absolute change in percentage of global DNA methylation in a population exposed to mandatory fortification of FA. No associations were found considering the other methyl donor nutrients.
Furthermore, in the present study, we did not find differences in the percentage of DNA methylation when comparing polymorphisms in folate metabolism genes. However, some studies have demonstrated DNA hypomethylation in individuals carrying the MTHFR 677 TT genotype(Reference Stern, Mason and Selhub41) and MTHFR 1298 AA genotype(Reference Friso, Girelli and Trabetti42). Other study explored DNA methylation pattern of the folate transporter genes FOLR1 (folate receptor 1), PCFT (proton-coupled folate transporter) and RFC (reduced folate carrier) in colorectal cancer. The results showed statistically significant differences in DNA-methylated fraction of all three genes at several gene regions(Reference Farkas, Befekadu and Hahn-Strömberg43).
Folate, being one of the methyl donor nutrients from one-carbon metabolism, has been extensively studied in cancer progression; however, there is no consensus on the relationship between folate and carcinogenesis(Reference Giovannucci11–Reference Kim, Smith-Warner and Spiegelman13, Reference Choi and Mason44). Nevertheless, there is an enormous interest in assessing the potential for changes in folate status to modulate DNA methylation as a mechanistic link to cancer development(Reference Crider, Yang and Berry9). Unfortunately, our study could not support these outcomes but a review analysed studies of cancer patients, folate status and global DNA methylation(Reference Crider, Yang and Berry9). In these studies, an association was found between cancer and global DNA hypomethylation in circulating blood and/or tumour tissue. However, no consistent association between global DNA methylation and folate status (intake or blood biomarker) across studies was found. So, the researchers concluded that the association of global hypomethylation with folate status (e.g. intake, blood folate concentration) has been inconsistent among studies of cancer patients.
One limitation of the present study was global DNA methylation analysis. There are several methods available to determine the DNA methylation in samples. However, it is important to note that all of the methods described in scientific literature have a tendency to either under- or overestimate the amount of global DNA methylation(Reference Kurdyukov and Bullock45). The correlation between these measures is actually quite low, and therefore, it is difficult to compare outcomes across studiesReference Wu, Delgado-Cruzata and Flom46.
In conclusion, no direct evidence was found in the present study that high dietary folate or FA intake or folate concentrations leads to DNA methylation. In this regard, the present study did not find an association between global DNA methylation and folate status (e.g. dietary intake and blood biomarkers) even in a population with a high FA intake due to mandatory flour fortification with FA, which has been in place since 2002. Only betaine intake, one of the methyl donor nutrients related to one-carbon metabolism, was associated with an absolute change in percentage of global DNA methylation. Clearly, further studies considering multidisciplinary approaches in epidemiology, genetics, biostatistics and bioinformatics are needed in order to understand the relationship between global DNA methylation and methyl donor precursors and their role as a marker of exposure and disease, specifically of cancer.
Acknowledgements
We thank the participants of the study and researchers from GEIAS (Grupo de Estudos Epidemiológicos e Inovação em Alimentação e Saúde) and GAC (Grupo de Pesquisa de Avaliação do Consumo Alimentar) and also the members (staff, technicians, students and scientists) of the Vitamin Metabolism Laboratory in the Jean Mayer United States Department of Agriculture Human Nutrition Research Center on Aging at Tufts University and Faculty of Pharmaceutical Sciences at University of São Paulo for helping during the biochemical analysis.
Financial Support from Municipal Health Secretariat of São Paulo, National Counsel of Technological-CNPq and Scientific Development and São Paulo Research Foundation – FAPESP (grant numbers: 2010/19899-5, 2011/19788-1, 2012/05505-0 and 2015/12196-2).
J. S., R. M. F. and D. M. M. conceived and designed the study. J. S., C. Z. P. and A. M. M. drafted and wrote the manuscript. J. S. analysed the data. J. S., C. Z. P., A. M. M., D. M. M. were responsible for critical analysis and final review. All authors read and approved the final manuscript.
The authors have no conflict of interest.







