Article contents
Estimating variance effect of QTL: an important prospect to increase the resolution power of interval mapping
Published online by Cambridge University Press: 14 April 2009
Summary
Equal variances within quantitative trait locus (QTL) groups in the segregating population are a usual simplifying assumption in QTL mapping. The objective of this paper is to demonstrate the advantages of taking into account potential variance effect of QTLs within the framework of standard interval mapping approach. Using backcross case as an example, we show that the resolution power of the analysis may be increased in the presence of variance effect, if the latter is allowed for in the model. For a putative QTL (say, A/a) one can compare two situations, (i) 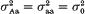 and (ii)
and (ii) 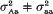 . It was found that, if the variance effect of A/a is large enough, then in spite of the necessity to evaluate an increased number of parameters, the more correctly specified model provides an increase in the resolution power, as compared to the situation (i). This is not unexpected, if either
. It was found that, if the variance effect of A/a is large enough, then in spite of the necessity to evaluate an increased number of parameters, the more correctly specified model provides an increase in the resolution power, as compared to the situation (i). This is not unexpected, if either 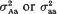 in (ii) is lower than
in (ii) is lower than 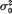 from (i). But our conclusion holds even if
from (i). But our conclusion holds even if 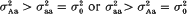 . These advantages are illustrated on sweet corn data data (F3 families of F2 genotypes). In particular, the log-likelihood test statistics and the parameter estimates obtained for a QT locus in the distal region of chromosome 2 show that the allele enhancing the trait is recessive over the opposite allele simultaneously for the mean value and variance.
. These advantages are illustrated on sweet corn data data (F3 families of F2 genotypes). In particular, the log-likelihood test statistics and the parameter estimates obtained for a QT locus in the distal region of chromosome 2 show that the allele enhancing the trait is recessive over the opposite allele simultaneously for the mean value and variance.
Information
- Type
- Research Article
- Information
- Copyright
- Copyright © Cambridge University Press 1996
References
- 13
- Cited by

