Introduction
The Canary Islands are characterized by a great diversity of endemic reptiles, which constitute a large part of the terrestrial vertebrate fauna of the archipelago (Arechavaleta et al., Reference Arechavaleta, Rodríguez, Zurita and García2010). Like other volcanic islands, the Canary Islands were colonized by reptiles through a series of long-distance dispersal events, in this case from mainland Africa (Illera et al., Reference Illera, Spurgin, Rodriguez-Exposito, Nogales and Rando2016). Three saurian genera, Gallotia, Chalcides and Tarentola, belonging to the families Lacertidae, Scincidae and Geckonidae, respectively, have diversified into 15 extant species via processes of adaptive radiation (Brown and Pestano, Reference Brown and Pestano1998; Carranza et al., Reference Carranza, Arnold, Mateo and López-Jurado2000; Cox et al., Reference Cox, Carranza and Brown2010).
Invasive species threaten biodiversity not only through predation or competition with native fauna, but also through the pathogens they can transmit. Emerging infectious diseases are an important phenomenon underlying biological invasions, and oceanic islands are more vulnerable to biological invasions than continental ecosystems (Gurevitch and Padilla, Reference Gurevitch and Padilla2004; Carroll, Reference Carroll2007). One invasive species in the Canary Islands stands out among the others and probably represents the greatest threat to the endemic vertebrates of the island of Gran Canaria: the California kingsnake, Lampropeltis californiae Blainville, 1835. The first observations of this North American colubrid were reported in southeastern Gran Canaria in 1998 (Pether and Mateo, Reference Pether and Mateo2007), but the eradication programme was not launched until 2007. Surveys in the following years revealed a large population that had expanded to the adjacent central and higher areas of the island, forming the so-called ‘main nucleus’ (NC1, Fig. 1). In 2010, the presence of a secondary nucleus (NC2) was confirmed in the northwest of the island (Gallo-Barneto et al., Reference Gallo-Barneto, Cabrera-Pérez, Peña-Estevez, Patiño-Martinez and Monzón-Argüello2016). Subsequent phenotypic and molecular studies suggested that the kingsnakes in these 2 nuclei originated from 2 different introductions (Monzón-Argüello et al., Reference Monzón-Argüello, Patiño-Martínez, Christiansen, Gallo-Barneto, Cabrera-Pérez, Peña-Estévez, López-Jurado and Lee2015). Despite the eradication programme, the establishment of a small population was also confirmed in the south of the island (the third nucleus, NC3) and in the northeast (the fourth nucleus, NC4). In all cases, populations of L. californiae occur in habitats that are considered Special Conservation Area (BOC no. 60, 15 May 2000) and are inhabited by a variety of endemic species.

Fig. 1. A map of Gran Canaria island showing 4 populations of Lampropeltis californiae as white-shaded areas (NC1–NC4); the place of collection of 10 snakes positive for Sarcocystis sp. based on coproscopic examination marked by red dots. NC1: Main nucleus. NC2: Secondary nucleus. NC3: Third nucleus. NC4: Fourth nucleus.
Dietary studies have confirmed the high proportion of endemic reptiles in the diet of snakes (69%), highlighting the Gran Canaria giant lizard Gallotia stehlini as the main component (43%), followed by the Gran Canaria skink Chalcides sexlineatus (23%) and the Boettger's wall gecko Tarentola boettgeri (3%) (Cabrera-Pérez et al., Reference Cabrera-Pérez, Gallo-Barneto, Esteve, Patiño-Martínez and López-Jurado2012; Monzón-Argüello et al., Reference Monzón-Argüello, Patiño-Martínez, Christiansen, Gallo-Barneto, Cabrera-Pérez, Peña-Estévez, López-Jurado and Lee2015). Small mammals (29.5%) and birds (1.5%) have also been detected in the digestive system of snakes. In addition to direct effects by predation on endemic fauna, possible co-invasive metazoan parasites infecting the snake have been detected, as well as ‘reverse’ infection with larval stages of parasites of feral cats and other vertebrates (Santana-Hernández et al., Reference Santana-Hernández, Orós, Priestnall, Monzón-Argüello and Rodríguez-Ponce2021).
Well-described evolutionary processes and the diversity of reptiles make the Canary Islands an interesting model site that has stimulated several parasitological studies involving both protists and helminths (Roca et al., Reference Roca, Jorge and Carretero2012; Jorge et al., Reference Jorge, Perera, Poulin, Roca and Carretero2018). Among protistan parasites with heteroxenous life cycles, 2 groups have recently gained attention – haemogregarines and Sarcocystis (e.g. Matuschka and Bannert, Reference Matuschka and Bannert1989; Bannert, Reference Bannert1992; Tomé et al., Reference Tomé, Pereira, Jorge, Carretero, Harris and Perera2018, Reference Tomé, Pereira, Harris, Carretero and Perera2019; Illera Cobo and Perera, Reference Illera Cobo and Perera2020). The latter genus is characterized by a 2-host life cycle, that typically involves predators and their prey (in the case of reptiles, commonly colubrid snakes and lacertid or geckonid lizards). However, Sarcocystis species described in Canary Island lizards of the genus Gallotia are particularly adapted to transmission in an environment where there are no predators. In their life cycle (usually referred to as dihomoxenous), merogony and gamogony occur in the same host species (in this case Gallotia lizards), which serves alternately as the final and intermediate host. Infection is transmitted among lizards via sporocysts in feces and cannibalism (Matuschka and Bannert, Reference Matuschka and Bannert1987, Reference Matuschka and Bannert1989).
The introduction of an invasive snake predator into a Macaronesian ecosystem inhabited by endemic saurian reptiles provides an unintended ‘experimental’ system to evaluate the host specificity of Sarcocystis spp. in, and its potential interactions with, the invasive kingsnake L. californiae.
Material and methods
Reptiles collected, examination
In the period 2019–2020, samples of California kingsnakes were analysed. The snakes were captured by the staff of Gestión y planeamiento territorial y ambiental (GesPlan) manually and using box-traps in the framework of the eradication project (https://www.gesplan.es/content/orden-33620-que-modifica-la-n%C2%BA-12419-ejecucion-plan-post-life-lampropeltis-y-actuaciones-del). The locality data, colour pattern, sex, length and weight of each animal (Table 1) were collected. Captured snakes were euthanized with intracardiac premedication of mixed ketamine and medetomidine (5–10 mg kg−1 and 0.15–0.5 mg kg−1), and a lethal dose (0.5–1 mL kg−1) of pentobarbital by the veterinarians from the eradication programme.
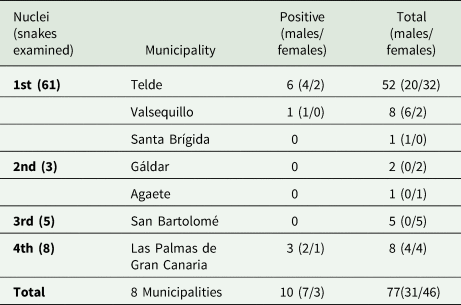
Table 1. Distribution of snakes regarding nuclei, the municipalities which encompasses and positive/negative to Sarcocystis sp.
The feces collected at the time of euthanasia was kept in sterile containers, transported in a refrigerator (3 °C) and examined within the following 12 h in the Parasitology Laboratory of the Faculty of Veterinary Sciences of the University of Las Palmas de Gran Canaria. The fecal samples were examined microscopically after centrifugal flotation with Sheather's sugar solution (Zajac and Conboy, Reference Zajac and Conboy2012), using a Nikon Eclipse Ni-U microscope with a Nikon Ds-Fi2 camera.
The small intestines of euthanized snakes were extracted and fixed in ethanol and 10% buffered formalin (Farris et al., Reference Farris, Squires, Ridgley, Lavergne, Serota and Mazzotti2013). After coproscopic examination, the small intestine samples that were positive for Sarcocystis sp. (i.e. in which sporocysts/ocysts were detected by microscopy) were further processed. Specimens preserved in formalin were processed for standard histopathological examination and stained with haematoxylin and eosin (H&E). Sections were examined with Olympus BX53 light microscope and photographed with Olympus DP73 camera and Olympus Dimension CellSens imaging software. As comparative material, 2 Gran Canaria giant lizards Gallotia stehlini Schenkel, 1901 were received dead from the Wildlife Recovery Centre of Tafira, 1 from the municipality of San Mateo and 1 from Arucas. The tail muscles were microscopically examined as squash preparations for the presence of Sarcocystis tissue cysts and preserved in ethanol.
DNA isolation, PCR and sequencing
A total of 7 snake fecal samples, 3 snake gut tissue samples, and 2 lizard tail muscle tissue samples, all stored in ethanol, were used for DNA isolation. Genomic DNA was isolated using the GeneAll Exgene™ Stool DNA mini kit for fecal samples and the NucleoSpin® Tissue kit (Macherey-Nagel) for tissue samples according to the manufacturer's instructions. The ssrRNA gene fragment was amplified using SarcoFext (5′-GGTGATTCATAGTAACCGAACG-3′)/SarcoRext (5′-GATTTCTCATAAGGTGCAGGAG-3′) primers (Moré et al., Reference Moré, Schares, Maksimov, Conraths, Venturini and Schares2013). To increase PCR sensitivity in isolates number 8 and 14, the following nested-PCR protocol was developed. A total volume of 25 μL PCR reaction mixture consisted of 12.5 μL PRCBIO Taq Mix Red Mastermix (PCR Biosystems Ltd.), 8.5 μL PCR H2O, 1 μL of each primer and 2 μL extracted DNA. Amplification began with an initial denaturation at 95 °C for 1 min, followed by 40 cycles of 95 °C (15 s), 57 °C (15 s), and 72 °C (15 s), and ended with a final elongation at 72 °C for 5 min. The primers used in the first run were SarcoFext/SarcoRext (see above) and in the second run SarcoFint (5′-CGCAAATTACCCAATCCTGA-3′)/SarcoRint (5′-ATCGTCTTCGAGCCCCTAAC-3′) (Moré et al., Reference Moré, Schares, Maksimov, Conraths, Venturini and Schares2013). PCR products positive in gel electrophoresis (1% agarose gel) were purified using the Gel/PCR DNA Fragments Extraction Kit (Geneaid Biotech Ltd.) and sequenced by Macrogen Europe B.V.
Phylogenetic and statistical analyses
The sequences obtained were processed using Geneious Prime® 2020.2.5 software (Biomatters Ltd.). The following sequences from GenBank (accession numbers in parentheses) were selected for comparison in phylogenetic analysis: Sarcocystis sp. (KX453662), S. lacertae (AY015113), S. gallotiae (AY015112), S. muris (KC877996), S. rodentifelis (AY015111), Frenkelia microti (AF009244), F. glareoli (AF009245), S. jamaicensis (KY994649), S. speeri (KT207459), S. neurona (U07812), S. ramphastosi (EU263366), S. falcatula (MH626537), S. lari (MF946588), S. rileyi (KJ396583), Sarcocystis sp. (KX833709) and Sarcocystis sp. (KX453661). Sarcocystis sp. (U97524) and S. atheridis (AF120114) as outgroup. Phylogenetic analysis was performed using the maximum likelihood method of the IQ-TREE web server tool (Nguyen et al., Reference Nguyen, Schmidt, von Haeseler and Minh2015) with ultrafast bootstrap (Minh et al., Reference Minh, Minh and von Haeseler2013). The phylogenetic tree was visualized and processed using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Basic statistical analysis was performed using SPSS v. 24.0 (IBM SPSS Corp., Chicago, Illinois, USA). The map was created using My map app by Google and GIMP 2.10 (GNU Image Manipulation program by Spencer Kimball, Peter Mattis and GIMP staff).
Results
A total of 77 kingsnakes and 2 lizards were examined for the presence of Sarcocystis sp. during the study period. Snakes from 7 different localities comprising the 4 nuclei were examined (Fig. 1). Seventy-eight per cent of the snakes were from the main nucleus. Only individuals from the main nucleus (Telde and Valsequillo) and the fourth nucleus (Las Palmas de Gran Canaria) were positive for the presence of Sarcocystis sp. in feces (Table 1). The average biometric parameters of the snakes studied are summarized in Table 2. A subtle difference was observed between positive (shorter and lighter) and negative animals.
Table 2. Biometrical parameters of positive and negative snakes to Sarcocystis sp.

All the measurements for length and weight are in millimetres and grams.
Coproscopy and histopathology
Oocysts and sporocysts of Sarcocystis sp. were detected by microscopy in the feces of 10 of the 77 snakes (Fig. 2). Liberated sporocysts (Fig. 2.1) were more common, although both types of exogenous stages were present in very low numbers. Sporocysts were broadly ellipsoidal, 11.5–12.5 × 9.7–10.3 (n = 5), with granular sporocyst residuum and 4 readily identifiable sporozoites.

Fig. 2. Developmetal stages of Sarcocystis sp. in feces after flotation (1–3) and in intestinal tissue (4 and 5) stained with H&E. 1: Isolated sporocyst with apparent sporocyst residuum; 2: intact oocyst with apparent oocyst wall; 3: oocysts deformed by preservation; 4: in situ sporulated oocyst with apparent sporozoites; 5 and 6: cross-section through oocyst in parasitophorous vacuole showing the sporocyst wall. All figures in same scale, scale bar = 20 μm, figs 3 and 5 photographed with Nomarski differential contrast.
Scattered developmental stages of gamogony of Sarcocystis sp. were observed in small intestinal sections of 3 California kingsnakes. The in situ sporulated oocysts were singly localized between enterocytes at the border of the lamina propria mucosae (Figs 2.4–2.6). No other developmental stages were seen.
Sequencing and phylogenetic analyses
Partial ssrRNA gene sequences of Sarcocystis sp. were obtained from 5 different California kingsnakes (2 from fecal sediment and 3 from small intestine tissue of snakes that were positive in histopathological sections) and from the tail muscle of a single Gran Canaria giant lizard. The sequences derived from the snakes were identical and 532–811 bp long (short sequences of 532 bp were from isolates processed by nested-PCR); the sequence from the tail muscle of the lizard was 785 bp long. The sequences obtained are deposited in GenBank under the following accession numbers: MW542198–MW542202 (from snakes) and MW542203 (from a lizard). The sequences obtained from the snakes and the lizard had 95.9% similarity. Phylogenetic analysis revealed that Sarcocystis sp. from the California kingsnakes in this study was most closely related to Sarcocystis sp. isolated from the crowned leaf-nosed snake (Lytorhynchus diadema – KX453661) and the common water monitor (Varanus salvator macromaculatus – KX833709), with pairwise similarity of 99.7% and 99.3%, respectively. Sarcocystis sp. isolated from the Gran Canaria giant lizard branched with S. gallotiae (AY015112) from G. galloti from Tenerife, with 99.7% similarity (Fig. 3).

Fig. 3. Phylogenetic analysis of the partial ssrDNA sequences showing position of the 2 taxa discussed. The sequences used for comparison were chosen from a phylogenetic tree containing all GenBank accessible ssrDNA sequences of Sarcocystis spp. (data not shown) and the current tree consists of the sequences most related to those extracted from L. californiae and G. stehlini, rooted on S. atheridis (AF120114) and Sarcocystis sp. (U97524). Geographical origin was added to each selected sequence.
Discussion
The finding of a Sarcocystis species in a population of invasive North American colubrid snakes recently introduced to Gran Canaria was unexpected, as the dixenous coccidia rarely invade new predator–prey systems. All the positive animals were found at the east nuclei of the island (main nucleus = 7/61 and fourth nucleus = 3/8). However, considering the huge difference of sample size between the nuclei no further statistical analyses were considered.
Three Sarcocystis spp. were previously described in endemic Gallotia lizards of Canary Islands, all of which have a dihomoxenous life cycle (Matuschka and Bannert, Reference Matuschka and Bannert1987, Reference Matuschka and Bannert1989; Bannert, Reference Bannert1992). In a previous phylogenetic study, Sarcocystis gallotiae was found to branch with Old World Sarcocystis species with a snake–lizard life cycle (Šlapeta et al., Reference Šlapeta, Modrý, Votýpka, Jirků, Koudela and Lukeš2001). Since Gallotia lizards evolved from North African ancestors (Cox et al., Reference Cox, Carranza and Brown2010; Illera et al., Reference Illera, Spurgin, Rodriguez-Exposito, Nogales and Rando2016), it is likely that the ancestral Sarcocystis had cycled between African lacertids and snakes. The DNA sequences of Sarcocystis that we obtained from invasive snakes and Sarcocystis from an endemic lizard G. stehlini differed significantly and branched into distant clades. On the contrary, the lizard-derived sequence was very close to S. gallotiae (>99 identity), which was previously isolated from G. galloti from Tenerife (Šlapeta et al., Reference Šlapeta, Modrý, Votýpka, Jirků, Koudela and Lukeš2001).
It is very likely that the Sarcocystis from the California kingsnakes described here is a snake-specific species that probably invaded the Macaronesian ecosystem along with its snake host. Sarcocystis species using reptiles as definitive hosts exhibit host specificity restricted to their host genus or family and are most likely unable to switch between hosts from different reptilian orders (Box and Smith, Reference Box and Smith1982; Lindsay et al., Reference Lindsay, Upton, Blagburn, Toiviokinnucan, Dubey, McAllister and Trauth1992).
Small mammals (such as rodents) are the most common intermediate hosts for Sarcocystis species that have a snake as a definitive host. However, some Sarcocystis spp. have been described to cycle between snakes and saurian reptilian hosts (Volf et al., Reference Volf, Modrý, Koudela and Šlapeta1999; Modrý et al., Reference Modrý, Koudela and Šlapeta2000). The intermediate host of Sarcocystis sp. found in the kingsnakes of Gran Canaria is still unknown and further research should be directed towards the study of native (snakes, skins, geckos, passerine birds) or invasive (mice and rats) vertebrates, which inhabit in all nuclei.
The exact origin of the presumably invasive Sarcocystis species remains unknown, as does the origin of the introduced California kingsnakes themselves. Maintenance of Sarcocystis infection in captive-bred snakes is unlikely because they cannot sustain a 2 host life cycles. On the contrary, the presence of patent Sarcocystis spp. infection in captive-bred snakes strongly suggests that they are wild-caught (Moré et al., Reference Moré, Pantchev, Herrmann, Vrhovec, Öfner, Conraths and Schares2014). In this sense, the presence of Sarcocystis sp. in invasive kingsnakes in Gran Canaria suggests that the population was established rather by individuals introduced from wild populations in the southwestern United States of northern Mexico than by captive-borne pet snakes.
Clarification of the life cycle of the snake sarcosporidian found in the Gran Canaria is necessary to answer questions related to possible impacts on the endangered endemic vertebrate fauna of Macaronesia.
Data
Data are available under reasonable requests.
Acknowledgements
The authors would like to thank the collaboration of Ramón Gallo Barneto, head of Gestión y planeamiento territorial y ambiental (GesPlan S.A.) as well as Miguel Ángel Cabrera Pérez, from Servicio de Biodiversidad, Dirección general de protección de la naturaleza, Gobierno de Canarias for donation of the specimens, to the personnel of GesPlan, who collected snakes in the field, the veterinary clinician in charge of euthanasia (Ayoze Melián) and Mr. de Blas for his help during the sampling and transportation.
Author contributions
David Modrý conceived and designed the study. All authors conducted data gathering and wrote the article.
Financial support
This study was supported by the project ‘Orden 336/20 que modifica la no. 124/19 ejecución plan post life + Lampropeltis y actuaciones del plan estratégico para el control de la culebra real de California (Lampropeltis californiae) en Canarias.’ financed by the government of Canary Islands and cabildo of Gran Canaria. https://www.gesplan.es/content/orden-33620-que-modifica-la-n%C2%BA-12419-ejecucion-plan-post-life-lampropeltis-y-actuaciones-del.
Conflict of interest
The authors declare there are no conflicts of interest.
Ethical standards
Not applicable.









