Introduction
Acetolactate synthase (ALS) (EC 2.2.1.6), also recognized as acetohydroxyacid synthase (AHAS; EC 4.1.3.18), catalyzes the formation of 2-acetolactate or 2-aceto-2-hydroxybutyrate in the biosynthesis of branched-chain amino acids (Duggleby et al. Reference Duggleby, McCourt and Guddat2008). It is well known that ALS is the target of ALS-inhibiting herbicides, which have been used worldwide since the early 1980s. Based on their different chemical structures, ALS-inhibiting herbicides are divided into the following classes: sulfonylurea (SU), pyrimidinyl-thiobenzoate (PTB), sulfonylamino-carbonyl-triazolinone (SCT), triazolopyrimidine (TP), and imidazolinone (IMI). To date, more than 160 weed species have evolved resistance to ALS-inhibiting herbicides worldwide due to continuous and intensive application (Heap Reference Heap2020). Resistance mutations in ALS isozymes that reduce the binding ability of ALS with herbicides are the most important and universal target-site resistance (TSR) mechanisms (Powles and Yu Reference Powles and Yu2010). To date, resistance mutations have been reported at the Ala-122, Pro-197, Ala-205, Asp-376, Arg-377, Trp-574, Ser-653, or Gly-654 loci of the ALS gene (corresponding to the sequence of ALS in Arabidopsis thaliana) (Heap Reference Heap2020; Yu and Powles Reference Yu and Powles2014).
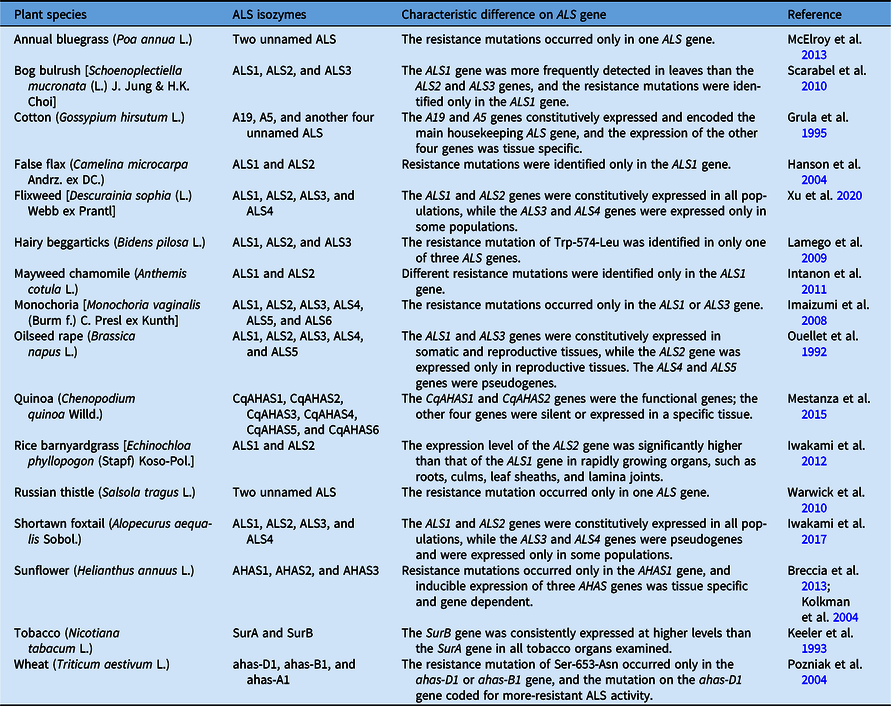
Multiple ALS isozymes have been reported in many plants (Table 1). Existing evidence suggests that different ALS isozymes might play different roles in the growth and development of plants. For example, the ALS1 and ALS3 genes are constitutively expressed in all somatic and reproductive tissues of oilseed rape (Brassica napus L.), while ALS2 transcripts have only been detected in flowers and young siliques and are regulated in an organ-specific manner (Ouellet et al. Reference Ouellet, Rutledge and Miki1992). Two ALS genes (SurA and SurB) have been detected in tobacco (Nicotiana tabacum L.), while the SurB gene is consistently expressed at higher levels than the SurA gene (Keeler et al. Reference Keeler, Sanders, Smith and Mazur1993). Similarly, two or more ALS isozymes were also found in various ALS-inhibiting herbicide-resistant weeds, and up to six ALS isozymes were found in quinoa (Chenopodium quinoa Willd.) (Mestanza et al. Reference Mestanza, Riegel, Silva and Vásquez2015) and monochoria [Monochoria vaginalis (Burm. f.) C. Presl ex Kunth] (Imaizumi et al. Reference Imaizumi, Wang, Ohsako and Tominaga2008) (Table 1). In resistant weeds, resistance-endowing mutations harbor only one of the ALS isozymes in a single plant, and the other isozyme is usually a wild-type copy. Although there are some reports for double resistance mutations in an individual plant, whether the mutations occur in the same or different target isozymes is still uncertain. However, in our previous studies, two ALS isozymes carrying resistance mutations were found in an individual tribenuron-methyl–resistant (TR) flixweed [Descurainia sophia (L.) Webb ex Prantl] (Deng et al. Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017). In the current study, a novel combination of two mutated ALS isozymes is identified in a single TR D. sophia plant.
Table 1. The information and characteristics of multiple acetolactate synthase (ALS) isozymes in plants, including weeds.
Descurainia sophia is a troublesome weed infesting winter wheat (Triticum aestivum L.) and has evolved resistance to tribenuron-methyl since its introduction into China in 1988. Amino acid substitutions in one or multiple ALS isozymes are responsible for D. sophia’s resistance to tribenuron-methyl. In addition, enhanced metabolism in TR D. sophia mediated by cytochrome P450s heightens D. sophia resistance (Deng et al. Reference Deng, Cao, Yang, Liu, Mei and Zheng2014, Reference Deng, Liu, Yang, Mei, Li and Zheng2015, Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017; Xu et al. Reference Xu, Xu, Li and Zheng2020; Yang et al. Reference Yang, Deng, Li, Yu, Bai and Zheng2016, 2018a, 2018b). In the current study, a new resistance mutation combination of mutated ALS1 (Asp-376-Glu) and ALS2 (Pro-197-Ala) is identified in individual D. sophia. Do weeds accumulate multiple mutated ALS enzymes in a single plant as a mechanism for combating higher herbicide selection pressure in fields? Are resistance mutations occurring on ALS isozymes randomly, or are some ALS isozymes are more preferentially mutated? Do different ALS isozymes play different roles in resistance evolution? Studies of these problems will help to reveal the resistance mechanisms to ALS-inhibiting herbicides. The current research aims to evaluate the contributions of different mutated ALS1 and ALS2 in resistance to different ALS-inhibiting herbicides in D. sophia through (1) determining the herbicide concentration that inhibits 50% of the activity of ALS1 and ALS2 (I50) and (2) comparing the expression levels of ALS1 and ALS2 genes in D. sophia.
Materials and Methods
Plant Materials
In 2016, seeds of the TR D. sophia population (SD1637) were collected from winter wheat fields in Liaocheng City, China (36.71°N, 115.84°E), where tribenuron-methyl had been used repeatedly for more than 20 yr. The tribenuron-methyl–susceptible (TS) D. sophia population (BJ1602) was harvested from the roadside in Beijing (40.21°N, 116.33°E), where tribenuron-methyl is unlikely to be used. To confirm the frequency of resistance mutations, the ALS genes for 50 plants in each population were sequenced, and all plants investigated had the same mutation combination.
Seeds of D. sophia were immersed in 20% H2O2 solution for 30 min, followed by a 24-h soak in 0.03% gibberellin solution, and germinated in petri dishes at room temperature for 4 d after rinsing with water. Sixteen germinated seeds were planted in one pot containing moist soil, which was kept in artificial climate chambers at 25/20 C (light/dark) for a 16-h photoperiod with a photosynthetic photon flux density of approximately 270 μmol m−2 s−1.
Determination of Resistance Mutations in ALS Genes
A DNA extraction kit (Plant Genomic DNA Kit®, DP305, Tiangen China, No. 86, Shuang Ying West Road, Changping District, Beijing, China) was used to extract genomic DNA from fresh leaves of TS and TR D. sophia. An F1/R1 primer pair was designed to amplify the ALS genes to full length (Table 2). The polymerase chain reaction (PCR) mixtures and program were the same as those described by Deng et al. (Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017). The PCR products were purified from agar gel with a purification kit (TIANgel Midi Purification Kit®, DP209, Tiangen China). The purified PCR products were ligated to the pLB-Simple vector and then transformed into TOP10 competent cells. Six clones of each DNA sample were selected for sequencing. Resistance mutations were identified by comparing the ALS sequence with that of susceptible D. sophia (accession no. JQ868736). The ALS genes for 50 plants in each population were sequenced to determine the proportion of plants carrying the resistance mutations.
Table 2. Information on primers for ALS gene cloning (F1/R1), reference gene 18S rRNA (F2/R2) and ALS expression determination (F3/R3, F4/R4) in tribenuron-methyl–susceptible (TS) and tribenuron-methyl–resistant (TR) Descurainia sophia.

a F, forward primer; R, reverse primer.
Whole-Plant Dose Response Experiments for ALS-inhibiting Herbicides
Whole-plant dose response experiments were used to determine the resistance or cross-resistance levels of TR and TS D. sophia populations to different ALS-inhibiting herbicides, including tribenuron-methyl (SU), flucetosulfuron (SU), pyribenzoxim (PTB), flucarbazone-sodium (SCT), flumetsulam (TP), and imazethapyr (IMI). Descurainia sophia plants at the 4-leaf stage were used for whole-plant dose response experiments. The herbicides were diluted to a series of concentrations with 0.2% Tween-80 solution. The spray doses used for ALS-inhibiting herbicides are listed in Table 3. Notably, due to the great difference in the susceptibility to tribenuron-methyl, the rates varied between TS and TR D. sophia populations. Herbicides were applied by using an automatic cabinet sprayer (ASP-1098 automatic sprayer, Zhejiang University Xinnong Pesticide Model Technology Development, Hangzhou, China) at a spray volume of 600 L ha−1. Control individuals were treated with water containing 0.2% Tween-80. The aboveground fresh D. sophia seedlings were weighed at 21 d after treatment. The experiments were conducted twice with three replicates in one dose.
Table 3. Herbicide rates of acetolactate synthase (ALS)-inhibiting herbicides in whole-plant dose response experiments and in vitro ALS activity assay.

a indicate the herbicide subfamily of sulfonylurea (SU), pyrimidinyl-thiobenzoate (PTB), sulfonylamino-carbonyl-triazolinone (SCT), triazolopyrimidine (TP), and imidazolinone (IMI), respectively.
b indicate the herbicide subfamily of sulfonylurea (SU), pyrimidinyl-thiobenzoate (PTB), sulfonylamino-carbonyl-triazolinone (SCT), triazolopyrimidine (TP), and imidazolinone (IMI), respectively.
c indicate the herbicide subfamily of sulfonylurea (SU), pyrimidinyl-thiobenzoate (PTB), sulfonylamino-carbonyl-triazolinone (SCT), triazolopyrimidine (TP), and imidazolinone (IMI), respectively.
d indicate the herbicide subfamily of sulfonylurea (SU), pyrimidinyl-thiobenzoate (PTB), sulfonylamino-carbonyl-triazolinone (SCT), triazolopyrimidine (TP), and imidazolinone (IMI), respectively.
e indicate the herbicide subfamily of sulfonylurea (SU), pyrimidinyl-thiobenzoate (PTB), sulfonylamino-carbonyl-triazolinone (SCT), triazolopyrimidine (TP), and imidazolinone (IMI), respectively.
f TS, tribenuron-methyl–susceptible; TR, tribenuron-methyl–resistant.
In Vitro ALS Extraction and Activity Assay
Approximately 40 d after planting, 4 g of leaf material was used for ALS extraction and activity assays in vitro according to the methods of Deng et al. (Reference Deng, Cao, Yang, Liu, Mei and Zheng2014). ALS activity in vitro was determined by colorimetry (520 nm) with a microplate photometer (Thermo Fisher, Waltham, MA, USA) by measuring acetoin production. The final concentrations in the reaction mixtures for the different herbicides are listed in Table 3. The experiments were repeated twice using independent enzyme extracts with three replicates for each herbicide concentration.
Relative Expression of ALS1 and ALS2 in TR and TS Descurainia sophia
Each D. sophia plant at 40 d after transplanting was treated evenly with 5 mg L−1 technical tribenuron-methyl (15 μl per plant) dissolved in a mixture of acetone and water (4:6 v/v) by using a micro-applicator (Hamilton PB 600 dispenser, Hamilton, Lancaster, PA, USA). The TS population exhibited no observable adverse response to the applied dose. Three plants were selected for RNA extraction by an RNA extraction kit (RNAprep pure Plant Kit®, DP432, Tiangen China) before tribenuron-methyl treatment (BT) or at 1, 3, 5, and 7 d after tribenuron-methyl treatment (DAT). Plants treated with only an equal volume of 40% acetone were used as controls.
18S rRNA was selected as a reference gene that was confirmed to be stably expressed in D. sophia (Yang et al. Reference Yang, Li, Shen, Xu, Liu, Deng, Li and Zheng2018b). Two primer pairs (F3/R3 and F4/R4) were designed for quantitative real-time polymerase chain reaction (qPCR) according to the differences between ALS1 and ALS2 gene sequences (Table 2). First-strand complementary DNA (cDNA) was synthetized according to the instructions of the FastQuant RT Kit (TIANScript II RT Kit®, KR107, Tiangen China). The expression level for ALS1 or ALS2 was determined by performing qPCR. The reaction mixtures with a volume of 20 μl consisted of 0.4 μl 50× ROX reference dye, 0.6 μl primers, 1 μl diluted cDNA, 7.4 μl ribonuclease-free distillation-distillation H2O (RNase-free ddH2O), and 10 μl 2× SuperReal PreMix Plus (SuperReal PreMix Plus®, FP205, Tiangen China). There were four replicates per cDNA. qPCR was carried out with programs of 15-min incubation at 95 C, 40 cycles at 95 C for 10 s, 60 C for 20 s, and 72 C for 32 s. There were four replicates per cDNA and three cDNA samples at each time point in the TS or TR populations.
Statistical Analysis
Whole-Plant Dose Response Experiments and In Vitro ALS Activity Assay
The GR50 (herbicide dose causing 50% plant growth reduction) and I50 were calculated using GraphPad Software (v. 5.0, San Diego, CA, USA) with Equation 1. This equation is based on a double-sigmoid model, which is constructed as the sum of two different logics. The double-sigmoid model suggests the coexistence of two inhibition targets with different susceptibilities.
In this equation, y is the percentage of fresh weight or ALS activity (% control); x is the log of the herbicide dose; and a and b are the logarithms of the GR50 (or I50) of two single-sigmoid curves (b > a). The Frac value is considered to be a putative proportion of the resistance contribution of more-susceptible ALS (Lipovetsky Reference Lipovetsky2010; Tsuneki et al. Reference Tsuneki, You, Toyooka, Kagawa, Kobayashi, Sasaoka, Nemoto, Kimura and Dani2004; Yamato et al. Reference Yamato, Sada and Ikeda2013).
Relative Expression of ALS1 and ALS2
The relative expression ratio (as 2−ΔΔC T) was calculated by the cycle threshold (CT) method (Schmittgen and Livak Reference Schmittgen and Livak2008), where ΔCT = CT target gene − CT internal control gene. Data for the relative expression level of ALS genes were tested for normality using the Kolmogorov-Smirnov test with SPSS software (v. 16.0, IBM, Armonk, NY, USA), and homogeneity of variance was confirmed by Levene’s test with SPSS. The data obtained from the relative expression of genes met the assumptions of an independent-samples t-test and ANOVA by testing. The relative expression of two ALS genes between TS or TR D. sophia was analyzed by the independent-samples t-test (P < 0.05). Dunnett’s test at the 5% level of significance in ANOVA was carried out to compare the relative expression of the ALS1 or ALS2 gene at different times in TS or TR D. sophia.
Results and Discussion
Determination of Resistance Mutations in ALS Genes
Two ALS genes with full lengths of 2,004 bp (ALS1) and 1,998 bp (ALS2) were cloned from TS (BJ1602) and TR (SD1637) D. sophia. A new combination of resistance mutations (ALS1 with Asp-376-Glu, ALS2 with Pro-197-Ala) was identified from a single TR D. sophia plant in the current study. The ALS1 and ALS2 genes in TS and TR D. sophia show high homology. A single different nucleotide caused the resistance mutation at the site of Pro-197 (in ALS2) or Asp-376 (in ALS1). Fifty TS or TR D. sophia plants were sequenced, and the ALS1 or ALS2 of each plant was the same in each D. sophia population, which indicated that the genetic backgrounds of the different plants were very similar or the same.
Two resistance mutations on ALS in a single plant were also reported in kochia [Bassia scoparia (L.) A.J. Scott; syn. Kochia scoparia (Linn.) Schrad] (Warwick et al. Reference Warwick, Xu, Sauder and Beckie2008), rigid ryegrass (Lolium rigidum Gaudin) (Kaundun et al. Reference Kaundun, Dale and Bailly2012), and Palmer amaranth (Amaranthus palmeri S. Watson) (Singh et al. Reference Singh, Singh, Salas-Perez, Bagavathiannan, Lawton-Rauh and Roma-Burgos2019), but it is not clear whether these mutations occur in the same ALS isozyme or in different ALS isozymes. To the best of our knowledge, a total of eight different amino acid mutations have been identified at Pro-197 (substituted by Ser, Thr, Leu, His, Ala, or Arg), Asp-376 (by Glu), and Trp-574 (by Leu) in ALS isozymes in TR D. sophia (Xu et al. Reference Xu, Xu, Li and Zheng2020). Initially, resistance mutations were only identified in one ALS isozyme, and the most common resistance mutation was identified at the site of Pro-197 (Xu et al. Reference Xu, Xu, Li and Zheng2020). However, resistance mutations at Asp-376 or Trp-574 in ALS, which can cause higher levels of resistance to tribenuron-methyl and broader cross-resistance to ALS-inhibiting herbicides than resistance mutations at Pro-197, were identified only in a few TR D. sophia (Deng et al. Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017; Xu et al. Reference Xu, Liu, Chen, Li, Liu, Wang, Fan, Wang and Ni2015; Yang et al. Reference Yang, Deng, Wang, Liu, Li, Yu and Zheng2018a). Deng et al. (Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017) first reported that two mutated ALS isozymes (Trp-574-Leu in ALS1, Pro-197-Thr in ALS2) in a single TR D. sophia plant exhibit a higher resistance level (789.3-fold) to tribenuron-methyl than TR D. sophia populations carrying a single mutated isozyme (211.8- and 366.3-fold) and exhibit cross-resistance to more ALS-inhibiting herbicides. In the current study, a novel combination of two mutated ALS isozymes (Asp-376-Glu in ALS1 and Pro-197-Ala in ALS2) was identified in a single TR D. sophia plant. Hence, we speculated that the accumulation of two or multiple mutated ALS isozymes in a single plant results in a higher level of resistance.
Whole-Plant Dose Response Experiments for ALS-inhibiting Herbicides
Whole-plant dose response experiments established the different sensitivities of TS (BJ1602) and TR (SD1637) D. sophia populations to different ALS-inhibiting herbicides (Table 4; Figure 1). The TS D. sophia were killed completely by tribenuron-methyl at a dose of 0.59 g ai ha−1, while all TR D. sophia survived at the highest dose of 150 g ai ha−1 (Figure 1A). The resistance index (RI) of TR D. sophia (SD1637) was 10,836.33, which is higher than that of the TR populations carrying any other known resistance mutations (Deng et al. Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017; Yang et al. Reference Yang, Deng, Wang, Liu, Li, Yu and Zheng2018a). For example, the populations with a single resistance mutation of Pro-197-His, Pro-197-Leu, Pro-197-Thr, Pro-197-Ser, Trp-574-Leu, or Asp-376-Glu in the ALS1 isozyme exhibited 205.8-, 250.0-, 708.4-, 263.7-, 485.3-, or 2,844.7-fold resistance to tribenuron-methyl, respectively (Yang et al. Reference Yang, Deng, Wang, Liu, Li, Yu and Zheng2018a), and the TR D. sophia population with two mutated ALS isozymes (Trp-574-Leu in ALS1, Pro-197-Thr in ALS2) evolved a 789.3-fold resistance to tribenuron-methyl (Deng et al. Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017). Therefore, the presence of two mutated ALS isozymes in individual plants helped weeds survive higher doses of ALS-inhibiting herbicide compared with weeds carrying a single mutated ALS isozyme. In addition, the accumulation of multiple ALS mutations in individual plants was also observed in A. palmeri (Singh et al. Reference Singh, Singh, Salas-Perez, Bagavathiannan, Lawton-Rauh and Roma-Burgos2019). In recent years, double mutations have been reported in a variety of resistant weeds, and these double mutations lead to weeds evolving higher resistance levels than weeds with a single resistance mutation. However, it is not clear whether double resistance mutations occur on the same or different target isozymes. For example, double mutations (Ala-251-Val and Phe-273-Val) in psbA resulted in Amaranthus spp. evolving a higher resistance to linuron and diuron compared with each single substitution (Davis et al. Reference Davis, Letarte, Grainger, Rajcan and Tardif2020). Goosegrass [Eleusine indica (L.) Gaertn.] with double mutations (Thr-102-Ile and Pro-106-Ser in 5-enolpyruvylshikimate-3-phosphate synthase [EPSPS]) developed a more than 32-fold higher resistance to glyphosate than E. indica carrying a single mutation (Pro-106-Ser) (Yu et al. Reference Yu, Jalaludin, Han, Chen, Sammons and Powles2015). Similar to E. indica, a different EPSPS double mutation (Thr-102-Ile and Pro106-Thr) conferring a high level of glyphosate resistance was reported in tetraploid beggarticks (Bidens pilosa L.) (Takano et al. Reference Takano, Fernandes, Adegas, Oliveira, Westra, Gaines and Dayan2020). Notably, a triple mutation (Thr-102-Ile, Ala-103-Val, and Pro-106-Ser) in EPSPS was reported, providing a 314-fold resistance to glyphosate compared with the wild-type strain in smooth pigweed (Amaranthus hybridus L.) (Perotti et al. Reference Perotti, Larran, Palmieri, Martinatto, Alvarez, Tuesca and Permingeat2018).
Table 4. The GR50 and I50 values of acetolactate synthase (ALS)-inhibiting herbicides for tribenuron-methyl–susceptible (TS) and tribenuron-methyl–resistant (TR) Descurainia sophia. a

a Abbreviations: GR50, herbicide rates causing 50% plant growth reduction; I50, herbicide rates inhibiting 50% ALS activity; RI, resistance index, GR50 value or I50 value of TR populations divided by that of TS population; NA, not applicable.
b Frac is considered to be a putative proportion of resistance contribution of more susceptible ALS.

Figure 1. Dose response curves of tribenuron-methyl–susceptible (TS, BJ1602-TS) and tribenuron-methyl–resistant (TR, SD1637-TR) Descurainia sophia treated with acetolactate synthase (ALS)-inhibiting herbicides: (A) tribenuron-methyl, (B) flucetosulfuron, (C) pyribenzoxim, (D) flucarbazone-sodium, (E) flumetsulam, and (F) imazethapyr. The experiments had three replicates per herbicide dose and were repeated twice. Vertical bars represent the standard error of two experiments.
The results obtained indicate that TR D. sophia exhibits obvious cross-resistance to representative herbicides of ALS-inhibiting herbicides with different chemical structures (Table 4). The cross-resistance levels to flucarbazone-sodium (SCT) and flucetosulfuron (SU) are much higher than to imazethapyr (IMI), flumetsulam (TP), and pyribenzoxim (PTB). Although the ALS isozymes are the common targets of these herbicides, their binding ability with ALS isozymes may be different due to different chemical structures. Therefore, TR D. sophia displays different cross-resistance levels to different ALS-inhibiting herbicides. The dose response of TR D. sophia to different ALS-inhibiting herbicides is different (Figure 1). The whole-plant dose response to tribenuron-methyl, flucetosulfuron, and pyribenzoxim can be fit to double-sigmoid models, demonstrating that two ALS isozymes in TR D. sophia have different sensitivities to these herbicides (Figure 1A–C). However, the curves fit for flucarbazone-sodium, flumetsulam, and imazethapyr are more consistent with single-sigmoid models, indicating that the ALS isozymes in TR D. sophia have similar sensitivities to these herbicides (Figure 1D–F). Frac is usually considered to be the proportion of resistance contributed by a more susceptible ALS isozyme to ALS-inhibiting herbicides (Yamato et al. Reference Yamato, Sada and Ikeda2013). The Frac value of TR D. sophia to tribenuron-methyl, flucetosulfuron, and pyribenzoxim indicates that the resistance contribution of the two ALS isozymes to these herbicides is obviously different. The resistance contributions to flucarbazone-sodium, imazethapyr, and flumetsulam are similar for the two ALS isozymes (Table 4).
In Vitro ALS Activity Assay
The results for I50 demonstrate that the herbicide concentrations inhibiting ALS activity in TR D. sophia increase greatly compared with those in TS D. sophia (Table 4). The curve-fitting models for the whole-plant dose response to and ALS activity inhibition by the same herbicide are nearly consistent for the TR population (Figures 1 and 2). The inhibition of ALS activity by tribenuron-methyl, flucetosulfuron, and pyribenzoxim can be fit by double-sigmoid models, indicating that two ALS isozymes in TR D. sophia contributed differently to the resistance to tribenuron-methyl, flucetosulfuron, and pyribenzoxim (Figure 2A–C). The ALS activity inhibition by flucarbazone-sodium, flumetsulam, and imazethapyr can be fit to single-sigmoid models, demonstrating that the two ALS isozymes exhibit similar contributions to the resistance to flucarbazone-sodium, flumetsulam, and imazethapyr (Figure 2D–F). One double-sigmoid curve can be regarded as the combination of two independent single-sigmoid curves. Compared with the single-sigmoid curves, the double-sigmoid curves for tribenuron-methyl, flucetosulfuron, and pyribenzoxim in TR D. sophia have a plateau between 1.0 to 10 μM, 10 to 100 μM, and 1.0 to 100 μM, respectively (Figure 2A–C), indicating that the more susceptible of the two ALS isozymes was saturated first. The activity of another ALS isozyme is gradually saturated with increasing herbicide concentration. In addition, the R2 obtained by weighted least-squares for the GR50 and I50 values for six ALS-inhibiting herbicides are determined to be 0.908, which indicates that the reduced ALS activity in response to herbicides is highly related to herbicide resistance in TR D. sophia. In particular, the I50-based RI value of flucarbazone-sodium is much lower than the GR50-based RI value. Given that the two ALS isozymes contribute equally to resistance to flucarbazone-sodium, the higher RI value (GR50-based) may also be due to non–target site resistance (NTSR) mechanisms. To date, NTSR to flucarbazone-sodium has been reported only in wild oat (Avena fatua L.) (Burns et al. Reference Burns, Keith, Talbert and Dyer2018). Further research is needed to determine whether the NTSR mechanisms confer flucarbazone-sodium resistance in TR D. sophia.

Figure 2. The acetolactate synthase (ALS) activity in vitro of tribenuron-methyl–susceptible (TS, BJ1602-TS) and tribenuron-methyl–resistant (TR, SD1637-TR) populations inhibited by ALS-inhibiting herbicides: (A) tribenuron-methyl, (B) flucetosulfuron, (C) pyribenzoxim, (D) flucarbazone-sodium, (E) flumetsulam, and (F) imazethapyr. The experiments had three replicates per herbicide concentration and were repeated twice. Vertical bars represent the standard error of two experiments.
Reduced ALS affinity with ALS-inhibiting herbicides, which is caused by resistance mutations in ALS isozymes, is the major TSR mechanism for D. sophia and other weed species (Deng et al. Reference Deng, Cao, Yang, Liu, Mei and Zheng2014, Reference Deng, Yang, Zhang, Jiao, Mei, Li and Zheng2017; Heap Reference Heap2020; Xu et al. Reference Xu, Liu, Chen, Li, Liu, Wang, Fan, Wang and Ni2015; Yang et al. Reference Yang, Deng, Wang, Liu, Li, Yu and Zheng2018a). However, most studies on the TSR mechanism for ALS have mainly focused on the sites and amino acids of resistance mutations in ALS isozymes. With the presence of multiple mutations in a single gene or multiple mutated target isozymes in an individual plant, the resistance patterns and mechanisms will become more complex. For example, the EPSPS crystal structure from Escherichia coli revealed that the mutation Thr-97-Ile (Thr-102 in plants) in the presence of Ser-101 (Ser-106 in plants) causes a shift of residue Gly-96 toward the glyphosate binding site, which impairs the efficient binding of glyphosate (Funke et al. Reference Funke, Yang, Han, Healy-Fried, Olesen, Becker and Schonbrunn2009). Compared with EPSPS with a double mutation, EPSPS with three mutations may form a new conformational structure with a smaller active site, leading to a higher exclusion of glyphosate (Perotti et al. Reference Perotti, Larran, Palmieri, Martinatto, Alvarez, Tuesca and Permingeat2018). The results here also indicate that the resistance contribution of two mutated ALS isozymes to tribenuron-methyl, flucetosulfuron, and pyribenzoxim is different, but there is no obvious difference in resistance to flucarbazone-sodium, flumetsulam, and imazethapyr (Table 4).
Relative Expression of ALS1 and ALS2 in TR and TS Descurainia sophia
The results in Figure 3 compare the relative expression levels of ALS1 and ALS2 in TS or TR D. sophia before treatment or at 1, 3, 5, and 7 DAT. At 1 and 7 DAT, the relative expression level of ALS1 in TR D. sophia is significantly higher than that in ALS2 by 2.2- and 1.6-fold, respectively. No obvious differences are observed before and at 3 and 5 DAT (Figure 3B). In contrast, the relative expression levels of ALS1 and ALS2 in TS D. sophia are not obviously different before or after tribenuron-methyl treatment (Figure 3A).

Figure 3. The relative expression of ALS1 and ALS2 genes in (A) tribenuron-methyl–susceptible (TS) or (B) tribenuron-methyl–resistant (TR) Descurainia sophia plants before tribenuron-methyl treatment (BT) or at 1, 3, 5, and 7 d after tribenuron-methyl treatment (DAT). Each data is the mean ± SE of four replicates. Means with different capital letters indicate the relative expression level of ALS1 was significantly higher than that of ALS2 at the same time point on level of P < 0.05. Means without letters show the relative expression level of two ALS genes at the same time point exhibited no significant difference.
The results in Figure 4 compare the relative expression levels of ALS1 (or ALS2) before or at 1, 3, 5, and 7 DAT in the same population. Compared with BT, the relative expression level of ALS1 in TR D. sophia is significantly increased 1.5- and 2.6-fold at 1 and 7 DAT (Figure 4A). ALS2 is not induced or slightly inhibited before 5 DAT and is increased 1.8-fold at 7 DAT (Figure 4B). In TS D. sophia, the relative expression level of ALS1 (or ALS2) at 1, 3, 5, and 7 DAT was approximately 0.7- (0.7-), 0.3- (0.4-), 0.4- (0.6-) and 0.6- (0.7-) fold that of BT, respectively (Figure 4).

Figure 4. The relative expression levels of ALS1 (A) (or ALS2, B) gene before tribenuron-methyl treatment (BT) or at 1, 3, 5, and 7 d after tribenuron-methyl treatment (DAT) in tribenuron-methyl–susceptible (TS) or (B) tribenuron-methyl–resistant (TR) Descurainia sophia. Each data is the mean ± SE of four replicates. Means with different capital (or lowercase) letters mean the relative expression level of ALS genes in TS (or TR) plants exhibited significant differences (P < 0.05) at BT or at 1, 3, 5, and 7 DAT, respectively.
The expression level of ALS1 is significantly higher than that of ALS2 in TR D. sophia, and ALS1 is more easily induced by tribenuron-methyl than ALS2. Therefore, ALS1 might be a preferred mutated target for weeds to gain resistance. The inducible expression of ALS has also been reported in other weeds. For example, the transcript levels for the ALS genes in B. scoparia were 7-fold higher 24 h after chlorsulfuron treatment (Varanasi et al. Reference Varanasi, Bayramov, Prasad and Jugulam2017). The transcript levels for AHAS3 in the roots of sunflower (Helianthus annuus L.) were induced 4- to 5-fold by imazapyr (Breccia et al. Reference Breccia, Vega, Felitti, Picardi and Nestares2013). ALS expression was upregulated 15.5- to 21.0-fold after mesosulfuron-methyl treatment in shortawn foxtail (Alopecurus aequalis Sobol.) (Zhao et al. Reference Zhao, Yan, Wang, Bai, Wang, Liu and Wang2018). Higher gene expression levels may lead to the accumulation of ALS isozymes, which can thus enhance the tolerance or resistance of weeds to herbicides by compensating for reduced ALS activity. However, enhanced transcription does not necessarily give rise to increased translation level, and posttranslational regulation may affect actual protein levels. Therefore, the contribution of increased ALS gene transcription to herbicide resistance needs more supporting data.
In summary, the novel resistance mutation combination (Asp-376-Glu in ALS1 and Pro-197-Ala in ALS2) leads to TR D. sophia evolving higher and broader resistance or cross-resistance to ALS-inhibiting herbicides than D. sophia with a single mutated ALS isozyme. The resistance contribution of two mutated ALS isozymes is different, but the contribution ratio of each ALS isozyme cannot be determined. The presence of two or multiple mutated ALS isozymes is probably the result of continuous herbicide selection on the basis of a single resistance mutation, which helps weeds survive higher doses of herbicides. However, it has to be pointed out that this study is based on a TS/TR D. sophia population. These results and speculations need to be verified by studying more D. sophia populations and even other weed species.
Acknowledgments
This work was sponsored by the National Natural Science Foundation of China (31672047). No conflicts of interest have been declared.










