Introduction
FINDER (www.finderc.org) is a new research project funded by the European Research Council (ERC) employing modern analytical techniques to address the lack of human fossils in the prehistoric record of Eurasia. The project's main goal is to identify and analyse new Denisovan, Neanderthal and early modern human remains from sites spanning the past 120 000 years. We apply peptide mass fingerprinting (also known as ZooMS—zooarchaeology by mass spectrometry; Buckley et al. Reference Buckley, Collins, Thomas-Oaies and Wilson2009) on large numbers of morphologically undiagnostic bone fragments to detect bones with characteristic Hominidae peptide markers. These are then analysed using ancient DNA, radiocarbon and U-series dating methods, which enable full genomic characterisation and age determination (Figure 1a).

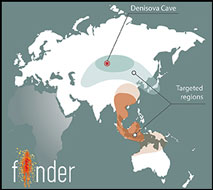
Figure 1. a) Schematic representation of FINDER's workflow; b) map of Eurasia with targeted regions of Late and Terminal Pleistocene sites.
Background
The Eurasian supercontinent is critical in understanding human evolution. Human fossils discovered in the past two decades (including those from Denisova Cave and Ust’-Ishim, Russia; Liang Bua and Lida Ajer, Indonesia; Tianyuan and Fuyan Caves, China; and Pestera çu Oase, Romania) have revealed much greater complexity in species spatio-temporal distribution and connections than previously envisaged. New discoveries of modern human remains suggest a substantially earlier presence in places previously lacking such evidence, while the geographic range of Neanderthals, traditionally considered a Europe-bound population, is now expanded significantly into Siberia and possibly northern China. We also know that Denisovans, an archaic sister group to Neanderthals, may have been distributed from Siberia to Melanesia over the past 200 000 years (for a recent review, see Bae et al. Reference Bae, Douka and Petraglia2017). To date, however, only five Denisovan remains have been discovered, all from the eponymous site (Douka et al. Reference Douka2019).
The significance of these findings encouraged us to address the noticeable lack of human remains from huge stretches of Eurasia. Understanding Pleistocene human presence and interaction “heavily depends on finding more Denisovan and other archaic human species remains” (Wolf & Akey Reference Wolf and Akey2018). Clearly, it is crucial that we develop means by which we can identify significantly more human bones from the archaeological record.
Materials, methods and preliminary remarks
Most Palaeolithic sites contain large numbers of bones that, due to post-depositional influences and human and carnivore processing, are either too small or lack the diagnostic features required for taxonomic identification. Our hypothesis is that these morphologically unidentified bulk assemblages probably contain parts of human skeletons too small to be identified as such. The use of ZooMS enables fast and cheap screening of hundreds of bones per week. Approximately 20mg of a sample undergoes a chemical pretreatment that results in the extraction and purification of collagen peptides (Figure 2), followed by spectrometric analyses using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-ToF-MS). Comparison of the spectra generated (Figure 3) against reference libraries of known taxa allows us to identify the unknown bones and separate faunal from hominin remains (Brown et al. Reference Brown, Higham, Slon, Pääbo, Meyer, Douka, Brock, Comeskey, Procopio, Shunkov, Derevianko and Buckley2016; Welker Reference Welker2018). Identifying human bones beyond the family level (Hominidae) is not possible using ZooMS; bone fragments with the characteristic peptide fingerprints therefore undergo DNA analysis for further genomic diagnosis. Direct radiocarbon and laser ablation U-series dating enables us to place the remains in their proper time period.

Figure 2. Illustration of the ZooMS methodology for the characterisation of unidentifiable bones. Each bone fragment (a) is sub-sampled and immersed in acid and base (b) to extract collagen (c). This is cleaved with trypsin (d) to create individual peptide sequences, which are spotted to a target plate (e) and measured in the MALDI-ToF-MS.

Figure 3. MALDI spectra showing taxonomically informative differences in the peptide fingerprints of different taxa (elephant, horse, bear and human). Peptide markers, defined previously for different taxa, are shown as individual peaks (A–G on the human spectrum).
We target two major geographic clusters stretching from Siberia to Papua New Guinea (Figure 1b), and expect to analyse around 40 000 bones. Since FINDER's launch in June 2017, we have collected, processed and analysed more than 5000 bones from the northernmost cluster of Palaeolithic sites, including Denisova and Strashnaya Caves in the Altai region of Siberia.
Future outlook
The discovery and scientific analysis of new human fossils will strengthen our ability to test interpretative models concerning the routes and timing of modern human movements across Eurasia, the frequency of interaction between indigenous and migratory populations, and, ultimately, the mechanisms leading to the demise and/or absorption of all archaic lineages and the sole global dominance of our species.
In addition to offering doctoral scholarships and ongoing volunteering opportunities, one of the main objectives of FINDER is training scientists and establishing new ZooMS preparation laboratories in the participating countries, where material can be processed locally. This will eventually enable archaeology departments and museums to design their own projects around the application of ZooMS, to expand the research questions to periods and materials not examined by FINDER, and to facilitate international collaboration in addressing pressing societal issues, ranging from anti-poaching efforts to effective communication of science in secondary education.
Acknowledgements
FINDER is funded by the ERC under the European Union's Horizon 2020 research and innovation programme, grant agreement 715069 to K.D. The DNA analyses are performed under the ERC project 100 Archaic Genomes, grant agreement 694707 to S.P., and the work of A.D. and M.S. is funded by the Russian Science Foundation (14-50-00036).