1. Introduction
In 1982, Victor McKusick, Physician-in-Chief of the Johns Hopkins University School of Medicine, published a commentary entitled, ‘The Human Genome Through the Eyes of a Clinical Geneticist’. At the time, McKusick was a central figure in the burgeoning field of medical genetics: a discipline populated by both MD physicians and PhD trained geneticists interested in the role of genetics in human disease.Footnote 1 Reflecting on the previous 25 years of progress in the field, McKusick noted,
The advances which started in 1956 have provided the clinical geneticist with his organ. Now the clinical geneticist is in the same position as the nephrologist with his kidney, the cardiologist with the heart, and so on. He has an organ that he can biopsy, of which he can analyse disordered structure and function, and which he can attempt to repair.Footnote 2
McKusick was pointing specifically to improvements in chromosomal observation, which, as the title of his paper suggests, helped MD geneticists to make a claim on their own ‘organ’: the human genome. At this time, during the early 1980s, McKusick and other medical geneticists were seeking to enhance the status and institutionalisation of their field within the medical profession. As part of this, they began pointing to the genome, as seen and represented by microscopically observable human chromosomes, as a distinct part of the human anatomy.
Historians of science and medicine have shown significant interest in the use of informational approaches in postwar genetics, which treat the human genome as an expansive digital data set composed of three billion DNA nucleotides.Footnote 3 Along these lines, scholars have focused on the use of various molecular techniques used to collect, decipher and analyse the DNA code.Footnote 4 Throughout the postwar period, however, geneticists have largely interacted with the genome at the microscopically visible level of physical objects: the human chromosomes. In this paper, I examine the observational and representational approaches of postwar human and medical geneticists. I argue that, during the 1970s and 1980s, new ways of seeing, standardising and depicting chromosomes began to reshape understandings of the human genome, leading to the broader recognition of it as a physical entity and a distinct part of the human anatomy. As I show, during the 1980s accounts of the human genome in the medical literature shifted away from discussing it as an informational abstraction composed of all of the human genes, and towards describing the genome as a discrete object, suitable for observational and clinical analysis.
Lorraine Daston has recently called for a ‘turn towards ontology’ among historians and philosophers of science, in particular, ‘towards ontologies created and sustained by scientific observation’.Footnote 5 Observation is a collective practice, argues Daston, and the building of visual ontologies is a gradual process, which takes place among a community that learns and agrees to see objects, both commonplace and obscure, in particular ways.Footnote 6 Along these lines, historians of science and medicine have begun to explore the observational practices of postwar human and medical genetics, in particular various approaches aimed at facilitating and standardising the microscopic observation of human chromosomes, and stabilising them as objects of study.Footnote 7
Unlike the abstract and sub-microscopic underpinnings of DNA code cracking and analysis, which had been previously highlighted by scholars, the study of chromosomes is highly visual, subjective, and fraught with ambiguous findings. Nonetheless, as Soraya de Chadarevian has noted, chromosomal analysis was of great value to human and medical geneticists, beginning in the 1950s, because it, for the first time, ‘offered a glimpse of the complete genetic make up on an individual’.Footnote 8 Indeed, this observational view of the human chromosomes produced extremely persuasive evidence, pointing to the discrete genetic basis of clinical disorders such as Down and Turner syndromes, while at the same time demonstrating that visible genetic abnormalities are often complex, variable and difficult to distinguish with absolute certainty.Footnote 9
Despite various complications, the ability to physically see the human chromosomes revolutionised the practices of postwar human and medical genetics. During the late 1950s and 1960s, the human chromosomes were slowly developed into important objects of study human and medical genetics. Susan Lindee has highlighted the important role of international standardisation meetings, held during the 1960s, in making the human chromosomes more scientifically and medically legible.Footnote 10 The clinical and research value of chromosomes was further extended in the next decade, following the introduction of new visual banding techniques. The impact of chromosomal banding on genetic research during this time has yet to be fully explored however, despite the central role of banded chromosomes in shaping how the human genome was depicted and communicated in the decades that followed.Footnote 11 In this paper, I trace the history of how standardised representations of banded human chromosomes came to be used by medical geneticists to depict the location of clinically relevant genetic traits in the genome.
In order to trace this observational and representational history of the human genome, I draw on published scientific and medical literature, medical textbooks, archival collections and interviews with human and medical geneticists. I begin with the introduction of chromosomal analysis to human genetics research, which offered geneticists their first ‘glimpse’ of the complete human genetic makeup during the 1950s and 1960s.Footnote 12 From here, I trace evolving depictions, definitions and conceptions of both the human chromosomes and the human genome in medical thinking, writing and public presentation during the 1970s and 1980s. In doing so, I demonstrate the ongoing process by which the human genome was reframed as an important object of medical analysis and understanding. As part of this, the genome was established as a visible and tangible entity, which influential medical geneticists claimed as their ‘organ’.Footnote 13
2. Counting Chromosomes in the 1950s
1953 is often pointed to as a landmark year in the history of genetics due to James Watson and Francis Crick’s proposal of a double helical structure for DNA. This important finding shaped the trajectory of genetics for years to come, because it led to an explanation of how DNA was replicated, and facilitated the ‘cracking’ of the DNA code, revealing the sequence-based correspondence between DNA nucleotides and the amino acids of protein. Over the next decade, these findings contributed to the growing informational orientation and basis of genetics research. The identification of the DNA double helix is regarded as an important moment in the history of medical genetics. However, Victor McKusick often pointed to another event, three years later, as being most important to the origin of the field, ‘medical genetics has become established as a clinical specialty, as the culmination of developments that began in 1956 with the description of the correct chromosome number in man’.Footnote 14
Until the early 1950s, the chromosome number in humans was believed to be 48.Footnote 15 The 1956 identification of the correct chromosome number, 46, is often pointed to as a significant moment in the history of medical genetics, because it led to the visual association of multiple clinical syndromes with abnormal numbers of chromosomes. Most notable among these were Down, Turner and Klinefelter syndromes, each of which were linked to specific chromosomal abnormalities by 1961. As McKusick put it in his 1997 history of medical genetics,
With the discovery of specific microscopically visible chromosomal changes associated with clinical disorders, beginning with Down syndrome in January 1959, medical genetics acquired an anatomic base. Medical geneticists now had their specific organ – the genome – just as cardiologists had the heart and neurologists had the nervous system.Footnote 16
It was at this time that the size and scope of the human genome, an increasingly important object of study in medical genetics, was concretely established and made relevant to clinical diagnosis.
Histories of medical genetics often highlight the 1956 demonstration, by Joe-Hin Tijo and Albert Levan, that humans normally possess 46 chromosomes.Footnote 17 As Aryn Martin has pointed out however, while ‘communal closure around the new number’Footnote 18 did not occur until 1956, the revolution in medical genetics associated with 1956 actually began with the introduction of a number of new techniques earlier in the decade. Foremost were the laboratory adoption of the chemical colchicine, which arrests cells at a point in the reproductive cycle when chromosomes are visible (they usually are not), and the use of a hypotonic (low-salt) solution to spread out a cell’s chromosomes, making them easier to see and count under the microscope.Footnote 19
As Martin suggests, the introduction of these new techniques began to change the way that geneticists thought about the human chromosome set, even before 1956. Referencing a 1952 paper by American cytogeneticist T.C. Hsu,Footnote 20 in whose lab the hypotonic technique was first (accidently) identified and used, Martin notes that Hsu, ‘also lined up the chromosomes in pairs by length and named them (by number) …[This] changed the counting game from the question “How many?” to the question “Are all members accounted for?”’Footnote 21 Establishing a singular ‘correct’ human chromosome number, therefore, was an important step in medical genetics towards physically bounding the size and scope of the human genome, allowing for a normal set of 46 human chromosomes to be distinguished from a pathological finding.Footnote 22
During the 1950s, human and medical geneticists developed new expectations for the counting of chromosomes. As a result, by the end of the decade, the number of chromosomes that an individual possessed had became much more meaningful, ‘A count of 47 is no longer indicative of a poor counting methodology, or a challenge to the established count, but of a body marked by genetic difference’.Footnote 23 Indeed, by the close of the 1950s, abnormal chromosomal counts were seen as very significant observational findings in both the laboratory and clinical contexts. Over the coming decades, the human chromosomes would be further transformed into standardised objects of biomedical thinking and practice, through the creation of various new observational and representational approaches in human and medical genetics.
3. Chromosomal Nomenclature in the 1960s and 1970s
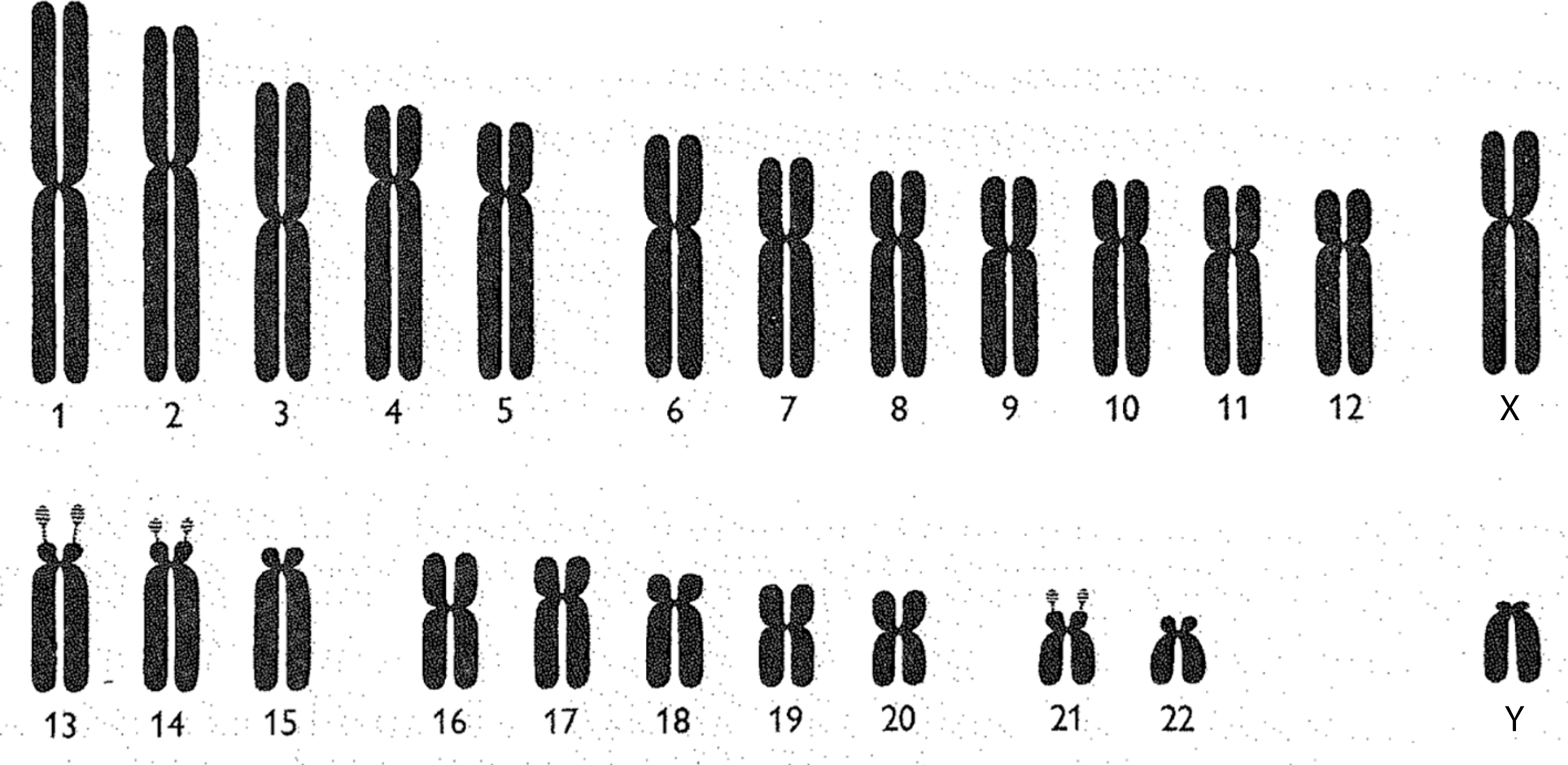
Chromosomes may be observed and represented in a variety of ways. One common depiction is the karyotype. When a karyotype is constructed, all of the chromosomes in a cell are photographed together, under a microscope, in one picture. Once this photograph has been developed, each chromosome is cut out and arranged by its relative size.Footnote 24 While karyotypes involve the cut-and-paste organisation of enlarged microscopic photographs, chromosomes may also be represented as idealised drawings called ideograms (figure 1). Chromosomal ideograms depict the distinctive features of each human chromosome. This includes arms, of which each chromosome has two (a long and a short arm), a centromere, separating the two arms, and satellites: additional material at the tip of the short arm of certain chromosomes.
Chromsomal ideograms are idealised depictions of the biologically ‘normal’. Ideograms, therefore, are not meant to capture exactly what a chromosome looks like under the microscope. Rather, ideograms provide the community of clinicians and geneticists who regularly observe chromosomes with standardised representations of each human chromosome’s distinctive features. When geneticists identify ‘abnormal’ chromosomes (showing the deletion, duplication or translocation of genetic material) during observational analysis, they often use ideograms to describe and map these ‘pathological’ findings. As I will argue in later sections, this use of standardised models of the ‘normal’ to depict the ‘pathological’, in medical genetics and more broadly, has been central to the development of contemporary biomedicine.Footnote 25

Figure 1: Human chromosomal ideograms developed by the 1960 Denver Study Group. From The Annals of Human Genetics 24: 319 (1960). Reprinted with permission from John Wiley and Sons.
In order to make chromosomes valuable scientific and clinical objects, geneticists created, and continuously revised, a standardised visual language for seeing, and communicating about, the anatomy of human chromosomes and any visible abnormalities. The development of a chromosomal nomenclature system was first undertaken at a 1960 international meeting of cytogeneticists in Denver. After much negotiation, the participants decided upon a nomenclature in which each ‘autosome’ (non-sex chromosome) was numbered by its relative size, with the largest being designated chromosome 1. The sex chromosomes continued to be called X and Y, even though the X chromosome is closer in size and shape to chromosome 7, and Y is similar to chromosome 22.Footnote 26 An international agreement to a specific numbering system was seen as quite important at this time, as more disorders were being associated with specific chromosomes.Footnote 27
While the Denver study group individually numbered each chromosome in 1960, it remained quite difficult, if not impossible, to microscopically distinguish the many chromosomes that were of similar size and shape. Reflecting this reality, geneticists also divided the human chromosomes into seven visually distinguishable groups: A (1–3), B (4, 5), C (6–12, X), D (13–15), E (16–18), F (19, 20), G (21, 22, Y). These groupings were officially recognised during a follow-up meeting of the Denver study group held in London in 1963. Within each lettered group, individual chromosomes were very difficult to tell apart. This situation was particularly problematic for the ‘C group’, which has eight members including the X chromosome.Footnote 28
The inability to differentiate individual chromosomes greatly limited the specificity of chromosome abnormality identification. For example, a medical geneticist could report that a member of the ‘B group’ was lacking its short arm in patients with a certain clinical disorder, but could not say for certain if the impacted chromosome was number 4 or 5. Later, another medical geneticist might find a similar aberration in a different group of patients, but not know whether the same chromosome was involved. As the 1960s went on, this difficulty in visually differentiating anatomically distinct human chromosomes greatly limited the ability of medical geneticists to associate clinical disorders with visible aberrations. In reference to this problem, McKusick, quoting his colleague Margery Shaw, referred to the late-1960s as an era when medical genetics found itself ‘in the doldrums’, due to inability to visually discriminate chromosomes.Footnote 29
The 1970s, on the other hand, proved to be another revolutionary time for the field, as new chromosomal banding techniques were introduced. The visibility of chromosomes had always been dependent on chemical staining, which greatly improved the contrast of chromosomes under the microscope. But, into the late 1960s, the available stains did little to help distinguish individual chromosomes. The introduction of fluorescent Quinacrine or ‘Q’ banding in 1968, and later Giemsa (G) banding and reverse (R) banding, in the early 1970s, changed this. These new staining techniques produced visually distinguishable banding patterns on each chromosome, allowing for them to be reliably differentiated by experienced observers.Footnote 30
The ability to concretely distinguish each chromosome was a significant contribution of new banding techniques, which became the basis for developing new, standardised approaches in human and medical genetics for the collective observation of chromosomes.Footnote 31 This, however, was only the beginning. Banding also created visible and reproducible landmarks on each chromosome, meaning that chromosomes could be broken down into additional regions and sub-regions based on their banding patterns. This advance was significant because it allowed human and medical geneticists to speak reliably in terms of much more than just the long or short arm of a certain chromosome. Now, a chromosomal aberration could be defined by its visually distinguishable and discrete ‘address’ along a particular chromosome.Footnote 32
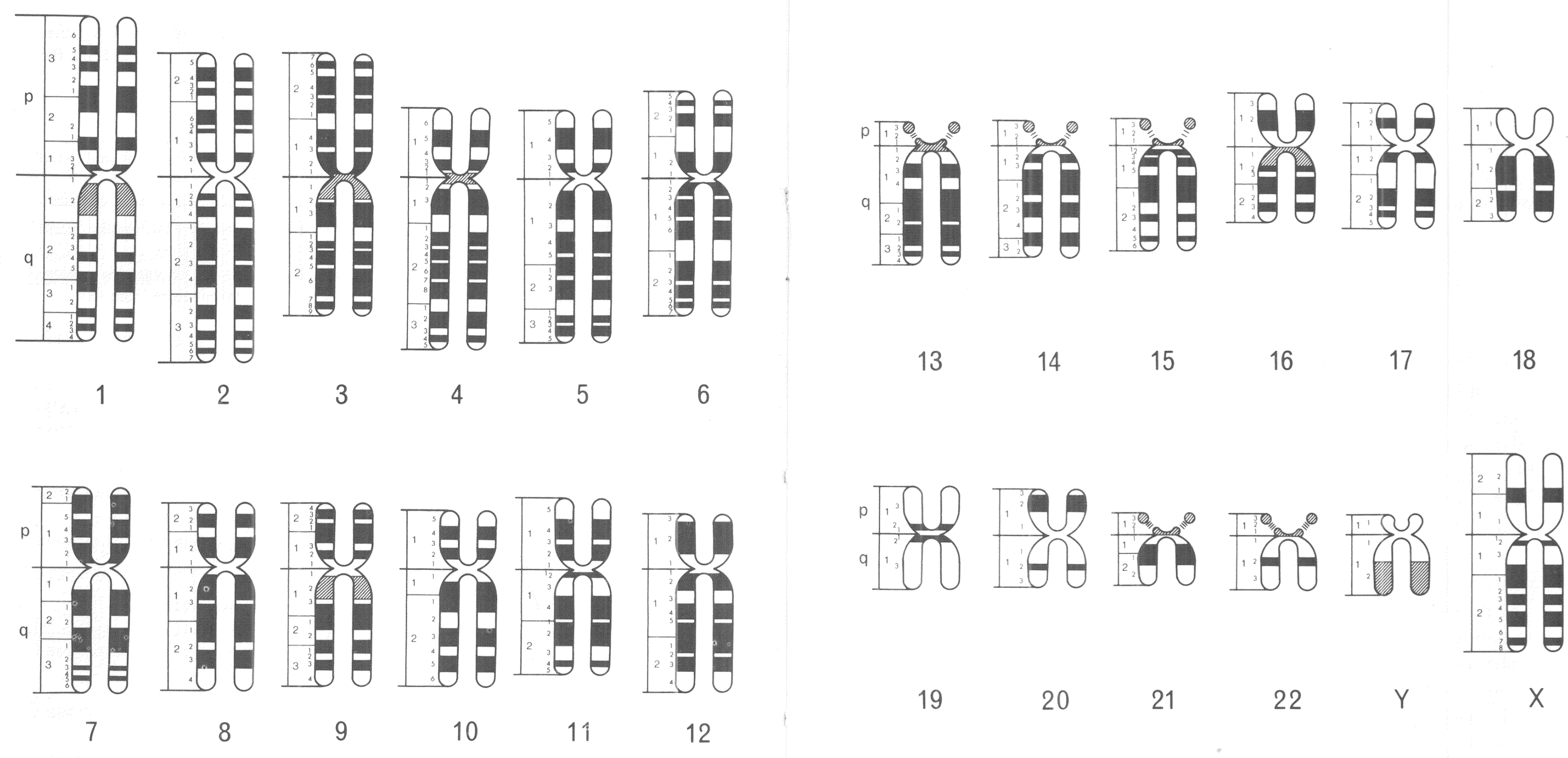
The standardised visual language of human chromosomes was soon after revised in order to incorporate this technological advance. An international committee was convened to do so in Paris during 1971. Following the numbering system established in 1960, members of the 1971 committee created a consensus nomenclature, which identified and systematically named consistently visible chromosomal bands. Chromosomal arms were broken down into anywhere from one to four regions, which were divided further by the bands visible in each. Bands were identified first by the chromosome number (1–22, X, Y), next by the chromosome arm (p or q), then with a number identifying the region (beginning with 1 at the centromere), and finally with a second number to identify the specific band (once again from the centromere).Footnote 33 Ideograms showing all of the (about 400) visible chromosomal bands among the 24 distinct human chromosomes (1–22, X, Y) were included in the 1971 conference report (figure 2).Footnote 34

Figure 2: Human chromosomal ideograms developed by the 1971 Paris Conference on Standardization in Human Genetics. Reprinted with permission from the March of Dimes.
This nomenclature was an important, and hard fought, international compromise, which provided a standardised visual language for human and medical geneticists. Different banding techniques (G and R) produced distinct visual results under the microscope, so coming up with one universal language proved challenging, especially since banding technique preferences broke down along international boundaries. Ultimately, however, variations in observational experience were set aside in order to facilitate the development of a standardised system for chromosomal representation.Footnote 35 This visual system of chromosomal nomenclature has been updated on many occasions, but has largely stood the test of time. These standardised depictions of banded chromosomes became central to the visual culture of human and medical genetics, in the 1970s and 1980s, and were used to represent growing knowledge about the human genome.
4. The Changing Look of the Human Chromosome Set
Since the first set of human chromosomal ideograms were proposed by the 1960 Denver Study group, eleven standardisation committee updates have been published, the most recent coming in 2013. Many of these revised editions suggested only small adjustments to the existing system, but a few provided a significant overhaul of what currently existed, most noticeably in the form of an entirely new set of chromosomal ideograms. Such major revisions occurred in 1971, 1981, 1995 and 2005. While each of these updates revealed the impact of innovations in chromosomal analysis, I argue that new sets of chromosomal ideograms also reflected, and offered, novel ways of seeing the thinking about the human genome. Here I focus specifically on various changes that were made to chromosomal ideogram depictions between 1970 and 1981.
The chromosomal ideograms developed by the 1960 Denver Study group have two distinguishing features when compared with those produced in later decades. First, the chromosomes are depicted as solidly coloured bodies: no banding techniques were yet available to visually provide each ideogram with a distinctive pattern. Second, each chromosome looks like some variation of a rounded-off letter X. The seven lettered groupings of human chromosomes, developed at a 1963 follow-up meeting in London, were based on variations in the chromosomes’ X-like shapes, as well as their relative size when observed under the microscope. Group A chromosomes are described as large and ‘metacentric’, meaning that their centromere is near the middle, and therefore that their long and short arms are close to the same size. Group B and C chromosomes are among those called ‘submetacentric’, since their long arms are significantly larger than their short arms. Group D and G chromosomes are ‘acrocentric’, meaning that their short arm is too small to easily be seen (figure 1).Footnote 36
In 1971, new chromosomal ideograms were developed, which were no longer solidly coloured, but instead were banded, reflecting the visual results of novel staining techniques. These banding patterns were, as I have already described, used for distinguishing each individual chromosome under the microscope. The 1971 ideograms also, notably, retained the rounded-off X-shape of previous versions (figures 1 and 2). Another fully new set of ideograms was not published until the next International System for Human Cytogenetic Nomenclature (ISCN) report in 1981. But, two intervening nomenclature updates, in 1975 and 1978, included human chromosomal ideograms that were no longer X-shaped, but rather looked like long, narrow rods. These ideograms were created, in part, to facilitate comparisons among the banding patterns of human and other primate chromosomes. The revisions were regarded as supplements to the 1971 Paris conference, as opposed to a full overhaul.Footnote 37 However, this new way of depicting chromosomes makes it clear that, in the mid-1970s, human and medical geneticists were beginning to think differently about what observational chromosomal analysis could reveal, and how standardised ideograms of human chromosomes could be used to represent these findings.
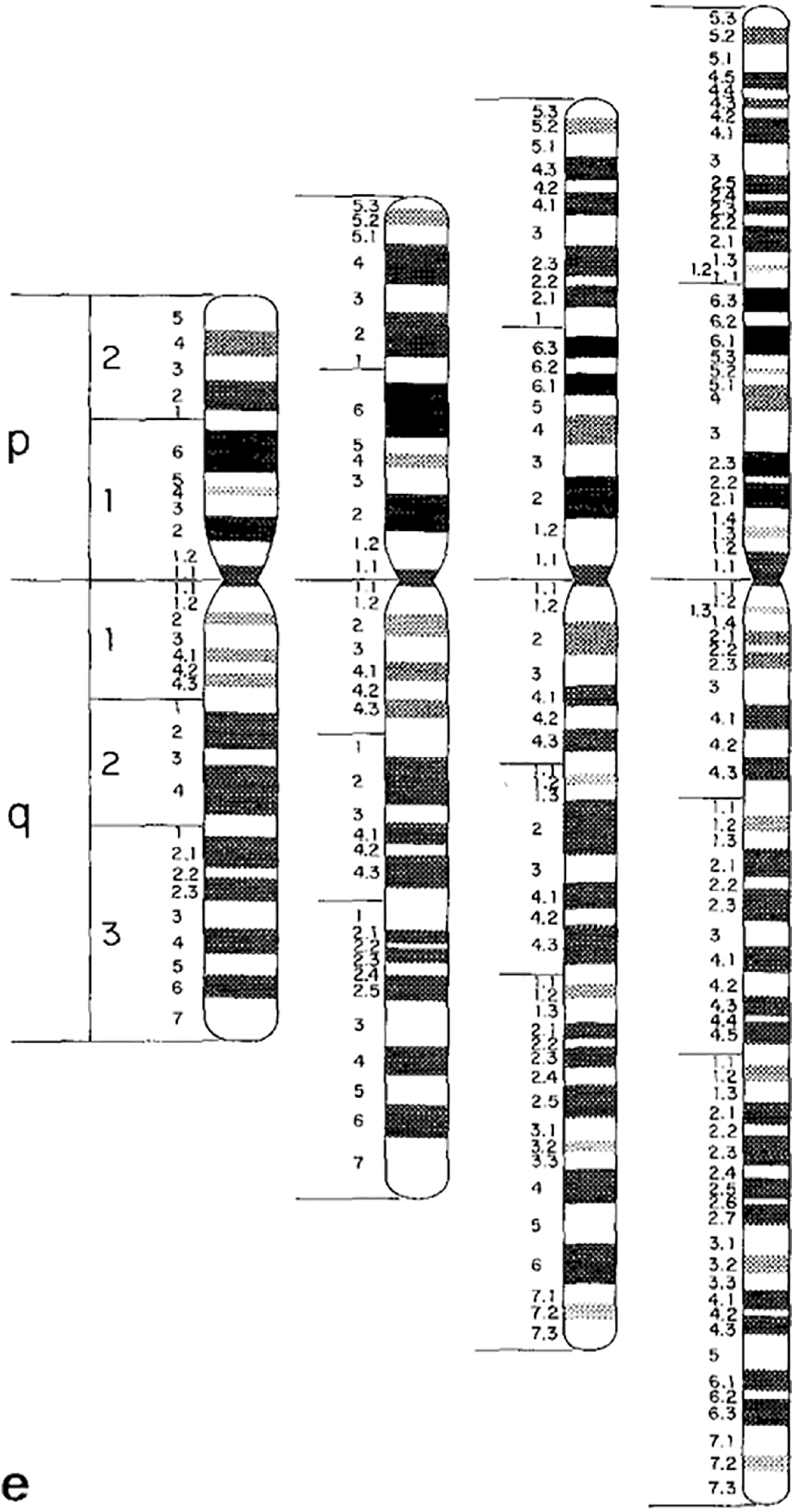
Undoubtedly, there were multiple reasons for this move from depicting chromosomal ideograms as X-shaped to rod shaped. Uta Francke, a German born medical geneticist who later moved to the US and was a consultant to the 1981 nomenclature conference, explained to me that part of the reason to switch from X-shaped to rod-shaped ideograms was efficiency: more ideograms could be fit on one page.Footnote 38 Another factor, in the late-1970s, was the introduction of high-resolution chromosome banding techniques.Footnote 39 High-resolution chromosomes are analysed earlier in the cell reproductive cycle, and as a result look more rod-like than X-shaped when observed under the microscope. A set of high-resolution chromosomal ideograms was included in the 1981 nomenclature update. These new ideograms featured a much denser banding pattern, totaling about 850 bands across the complete set of human chromosomes (figure 3).Footnote 40

Figure 3: Banded and rod shaped chromosomal ideogram, based on high-resolution analysis. Figure 3e from: J.J. Yunis, J.R. Sawyer and D.W. Ball, ‘The Characterization of High-resolution G-banded Chromosomes of Man’. Chromosoma 67, 4 (1978), 293–307: 301. Reprinted with kind permission from Springer Science and Business Media.
X-shaped chromosomal ideograms had, as they were depicted prior to 1975, represented important functional units of cellular reproduction. These ideograms were shown to be X-shaped because they actually represented two exact copies of the same chromosome (joined at the centromere), which were about to split apart in the formation of two genetically identical cells. Chromosomal analysis up to this point was mostly focused on identifying large chromosomal abnormalities. With the wider uptake of chromosomal banding in the early-to-mid-1970s however, the capabilities and the aims of chromosomal analysis began to shift. Indeed, it was becoming possible to identify and communicate about much more discrete locations than just entire chromosomes, or large portions of them. As a result, the human chromosomes were coming to be understood as more than just units of cellular reproduction: they were also increasingly seen as the basic, observable and mappable subsections of the human genome.
Banded chromosomal analysis facilitated early attempts, during the 1970s, to produce a ‘physical map’ of the human genome. Physical mapping involved the association of genetic characteristics with distinct genomic landmarks. This process was different from the older, but still important, technique of ‘linkage’ or ‘genetic’ mapping, famously practiced in T.H. Morgan’s fly lab. Linkage mapping sought to identify the relative location of genes, to one another, on individual chromosomes. In physical mapping, on the other hand, geneticists worked to associate genetic traits with fixed genomic locations. During the 1970s and 1980s these landmarks were most often based upon microscopically observable chromosomal bands.Footnote 41
Physically speaking, and from a mapping perspective, the human genome was conceived of, at this time, as a linear chain of DNA. As the medical genetics textbook The Metabolic Basis of Inherited Disease put it in 1978, ‘All genetic mapping data are consistent with the hypothesis that the genome is a linear unbranched structure’.Footnote 42 Genes were thus understood to be linearly arranged, one after another, along a strand of DNA. With this conception of the human genome in mind, therefore, it would not have made visual, or logical, sense to map genes onto X-shaped chromosomes, which as, depicted before 1975, contained four physically discontinuous arms.
This said, there was no indication in the cytogenetic standardisation committee reports from 1975, 1978 or 1981 that the shift from X-shaped to rod-like chromosomes was meant to make the human genome more mappable,Footnote 43 and it is not my intention to argue for a causal link in either direction between ideogram linearity and genome mappability. Rather, I suggest that this alteration in chromosomal representation during the mid-1970s reflects a shift in how human and medical geneticists conceptualised the relationship between the assumed linearity of the human genome and the microscopically observed characteristics of human chromosomes. In the next section, I demonstrate this same shift from another perspective: by tracing the changing use and definition of the term ‘genome’ in medical genetics and general medical textbooks.
5. Evolving Conceptions of the Human Genome
It is commonly held that German botanist Hans Winkler coined the term ‘genome’ in 1920 as a hybrid of the words ‘gene’ and ‘chromosome’.Footnote 44 However, an alternative interpretation, offered by Joshua Lederberg and Alexa McCray in 2001, holds that the suffix ‘ome’, used by Winkler in 1920 instead referred to, ‘a holistic abstraction, an eventual goal, of which only a few parts may be initially at hand’, as in the use of ‘biome’ to refer, in a general sense, to all living things in a particular Earth environment.Footnote 45 In this section, I trace the meaning of the term ‘genome’ in medical textbooks from the late-1960s through the 1980s. My findings show a shift in the use of ‘genome’: from being an abstract way to identify all of an individual’s genetic material or genes, to referring to a observable and physically bounded anatomical entity.
A number of textbooks aimed at geneticists, interested in studying the genetic basis of human disease, were in print during the 1970s. ‘Genome’ only appears in a couple of these textbooks, and when it does, it is described in quite abstract terms. The glossary of McKusick’s text Human Genetics (1969), defines the genome quite simply as ‘The total genetic endowment’.Footnote 46 While the 1973 edition of Genetics in Medicine, a textbook by physician James S. Thomas and PhD geneticist Margaret W. Thompson, defines genome in its glossary as ‘The full set of genes’.Footnote 47 Beyond this, the term genome does not appear in the index or the main text of other medical genetics textbooks available during the 1970s, such as An Introduction to Medical Genetics (1973) and Medical Genetics: Principles and Practice (1974).Footnote 48
Discussions and definitions of the human genome were equally absent in more general medical texts during the 1970s. The term genome is not used in either the 1971 or 1975 editions of the Cecil Textbook of Medicine.Footnote 49 In the 1979 update, genome is not listed in the textbook’s index. However, in a chapter on genetics, physician Alexander G. Bearn does muse, quite abstractly, ‘It is apparent that despite the acceleration in discovery of new genetic entities 90 per cent of the human genome remains to be discovered’.Footnote 50 Genome does, in fact, appear in the 1970 and 1974 editions of another prominent general medical text, Harrison’s Principles of Internal Medicine. The term can be found in a chapter contributed by McKusick, which refers to the genome in 1970 as ‘the rest of the genetic make-up’, and as ‘the genetic background’ in 1974.Footnote 51 Following the departure of McKusick as the author of this chapter, however, the term genome completely disappears from the 1977 and 1983 editions of Principles of Internal Medicine, in which physicians Joseph L. Goldstein and Michael S. Brown contributed a similar chapter on genetics and disease.Footnote 52
During the mid-1980s, the term genome became both more prominent and more precisely defined in medical genetics and general medical textbooks. The 1986 edition of Thompson and Thompson’s Genetics in Medicine states ‘The term genome refers to the full DNA content of the chromosome set’. An Introduction to Medical Genetics (1985) by physicians J.A. Fraser Roberts and Marcus Pembrey refers to genomic DNA as ‘the nuclear DNA of the chromosomes’. Physician James J. Nora and PhD geneticist F. Clarke Fraser define genome as ‘The complement of genes found in a set of chromosomes’ in the 1981 edition of Medical Genetics: Principles and Practice.Footnote 53 In the 1985 update of the Cecil Textbook of Medicine, contributor John L. Hamerton, a human geneticist, notes ‘The term genome refers to the full DNA content of the chromosome set’. Genome also appears once again in the 1987 edition of Harrison’s Principles of Internal Medicine, in which physician and cell biologist Arthur Beaudet compares the human genome to a series of books which ‘can be envisioned as being bound into 46 volumes, each the equivalent of one chromosome’.Footnote 54
It would be inaccurate to suggest that the genome had not been associated with the human chromosomal complement before 1980. However, entries from these medical textbooks clearly show that there was a shift in the importance and meaning of the term genome for medical geneticists, and physicians more broadly, between the early 1970s and mid-1980s. When used in the 1970s, genome generally referred to the abstract concept of ‘all the genetic material or genes’ possessed by an individual. By the mid-1980s, however, as the term became increasingly commonplace, the genome was more often defined in the context of the human chromosomes. Indeed, during the 1980s, the human genome came to be understood as a discrete object of interest and examination among human geneticists, as well as more clinically oriented medical geneticists. As part of this, the genome was widely represented as being physically embodied by a microscopically observable part of the human anatomy: the chromosomes.
6. The Human Genome as an Anatomical Entity
As I have already described, beginning in the early 1980s, McKusick often spoke of the human genome (and alternatively of the human chromosomes) as being the ‘organ’ of medical genetics, equating it with the heart for cardiologists and the kidneys for nephrologists. This concept fit into his larger campaign, which presented the human genome as an important part of the human anatomy.Footnote 55 McKusick cited multiple sources for this conception of the genome, including the influential human geneticist Curt Stern and McGill medical geneticist Charles Scriver.Footnote 56 Clearly, McKusick found the metaphor to be of great descriptive and rhetorical value, as he used it in most every paper he published on the genome from 1980, until his death in 2008.
McKusick was not alone in using anatomical analogies to speak about the human genome during the early 1980s. Molecular biologist Paul Berg, in his 1980 Nobel Prize lecture, made a similar anatomical reference when talking about the genome and its relevance to medicine,
Just as our present knowledge and practice of medicine relies on a sophisticated knowledge of human anatomy, physiology, and biochemistry, so will dealing with disease in the future demand a detailed understanding of the molecular anatomy, physiology, and biochemistry of the human genome …. We shall also need physicians who are as conversant with the anatomy and physiology of chromosomes and genes as the cardiac surgeon is with the structure of the heart and the circulatory tree.Footnote 57
Scriver also adopted an anatomical metaphor for speaking about the human genome in the early 1980s, when referring to genomic mapping as being akin to a ‘neo-Vesalian anatomy’.Footnote 58 McKusick picked up in this exact phrasing soon thereafter (at first without citing Scriver), noting in 1986 that, ‘Knowledge of the chromosomal and genic anatomy of Homo sapiens has given clinical genetics (and medicine as a whole) a neo-Vesalian basis’.Footnote 59 In his account of the early history of the Human Genome Project, Gene Wars (1994), Robert Cook-Deegan recounts that such references to the ‘neo-Vesalian’ nature of genome mapping were quite successful in attracting funding sources for these projects, in particular from the Howard Hughes Medical Institute.Footnote 60 Indeed, these historical and anatomical references made mapping the human genome appear more legible and relevant to a wider audience of both physicians and research funders.
When pointing to the ‘neo-Vesalian’ basis of medical genetics, McKusick and Scriver were referencing the work of Andreas Vesalius, a sixteenth century physician, famous for his anatomical images published in On the Fabric of the Human Body (1543). The frontispiece of this text, which McKusick published as part of his 1981 paper, ‘The Human Genome Through the Eyes of Mercator and Vesalius’, depicts Vesalius teaching human anatomy by directly pointing to a newly dissected human body. This was in contrast to the existing norm of the sixteenth century, when an instructor would read directly from the 1400-year-old text of Galen while standing apart from the dissected body.Footnote 61
To the present day, Vesalius is remembered in the western medical community as having brought direct observation of the dissected human body back to the forefront of research and teaching in human anatomy. In The Secrets of Women: Gender, Generation, and the Origins of Human Dissection (2006), however, Katherine Park counts Vesalius among many fifteenth and sixteenth century figures who have been inaccurately remembered as heroes because they ‘braved persecution and censure in the service of art and science’. As Park suggests, continued reference to these figures does ‘important cultural work’, providing ‘foundation stories that confirm deep-seated Western institutions about the scientific origins of modernity – institutions that continue to inform the writing of even specialists in the field’.Footnote 62
Indeed, late twentieth century physicians such as McKusick and Scriver regarded Vesalius as a revolutionary figure, who had an impact on future centuries of medical thinking and practice. References to the ‘neo-Vesalian’ nature of late-twentieth century medical genetics were rhetorically valuable for making the argument that the genome mattered to medical practice because it was physically and visually a part of the human anatomy. In addition, just as the work of Vesalius was seen as reshaping medicine in the sixteenth century and beyond, during the 1980s, anatomical exploration of the human genome was similarly presented by McKusick and Scriver as likely to have revolutionary implications for medicine in decades to come.
Scriver now refers to the gene mapping, which was taking place during the 1970s and 1980s, as ‘the genome project of the day’. As a member of the Howard Hughes Medical Institute (HHMI) advisory board during the 1980s, Scriver was a major proponent of funding this ongoing research. Scriver described his interest in gene mapping at the time to me in this way, ‘There was lots of initial episodic work, where a certain gene might be mapped to a certain particular region of a chromosome, and so a mosaic was being built up. I was interested in seeing the whole picture being completed’.Footnote 63 With funding from the HHMI, Scriver hoped that this vast collage of genetic entities, which made up the human genome, could one day be fully catalogued. As I describe in the next section, during the late 1970s and 1980s, McKusick used representations of chromosomal anatomy to layout the conceptual framework for doing just this.
7. The ‘Morbid Anatomy’ of the Human Genome
In addressing the mapping of human genetic traits as part of his paper, ‘The Human Genome Through the Eyes of Mercator and Vesalius’, McKusick noted, ‘for an ever increasing number of diseases the chromosomal location of the mutant gene responsible is known’.Footnote 64 Similar to the ways in which diseases may be located in the bodies of patients, their etiological cause may also be located in the human genome. McKusick referred to this practice as looking at the ‘morbid anatomy’ of the human genome, citing the 18th century work of Giovanni Morgagni in locating the ‘clinical pathology’ of a disease in certain bodily organs.Footnote 65 Scriver similarly made reference to this way of thinking about the human genome in his 1982 paper, ‘Window Panes of Eternity. Health, Disease, and Inherited Risk’, noting, once again in reference to Vesalius’ 1543 anatomy text, ‘Another revolution in anatomy is occurring; it is chromosomal and genetic cartography achieved by mapping of genes to specific chromosomes and bands on chromosomes and the delineation of nucleotide sequences in specific genes, respectively. We are beginning to possess chromosomal addresses for Mendelian disease’.Footnote 66 Indeed, in the early 1980s, the human genome was being looked to as a place within the human body where the discrete causes of genetic disease could be seen, mapped and potentially understood.
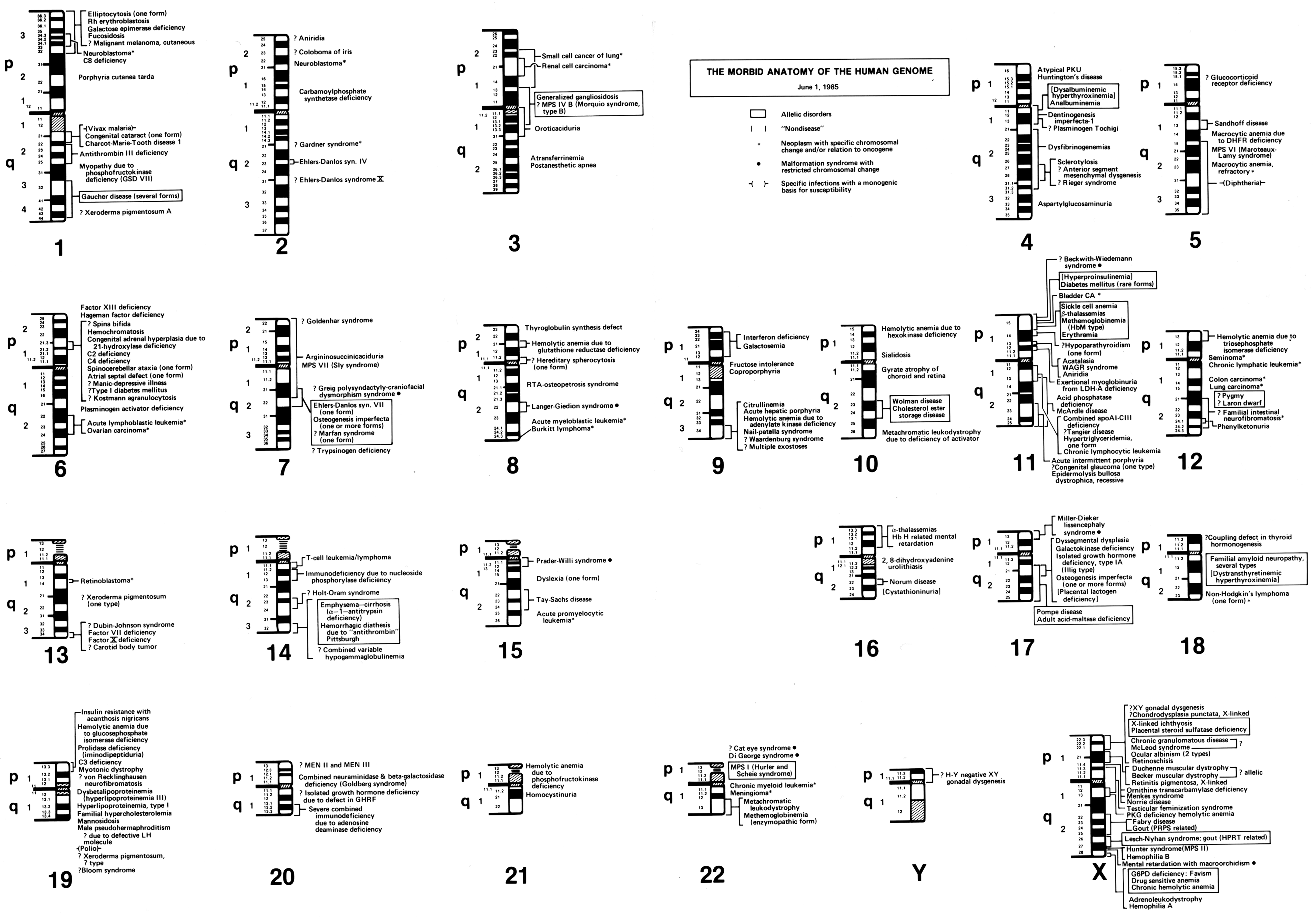
In his 1982 paper, ‘The Human Genome Through the Eyes of a Clinical Geneticist’, McKusick published what he called ‘The Morbid Anatomy of the Human Genome’. To illustrate this ‘morbid anatomy’, McKusick started with the 23 sets of standardised human chromosomal ideograms (1–22, XY), which showed the newly revised banding patterns from the 1981 Paris Conference on nomenclature. Along each chromosome, genetic diseases that had been mapped to certain genomic locations were identified. Some disorders were known only to be associated with a specific chromosome, while others were linked more specifically to a particular chromosomal region or band (figure 4).Footnote 67 This was not the first time that McKusick had created such a figure mapping genetic elements along the human chromosomes. In 1977, McKusick, along with Yale University geneticist Frank Ruddle, published a similarly structured human gene map, which was created to celebrate a recently achieved milestone: by the end of 1976, at least one gene had been mapped to every human chromosome.Footnote 68

Figure 4: Victor McKusick’s ‘The Morbid Anatomy of the Human Genome’ figure from 1 June 1985. Reprinted with permission from The Alan Mason Chesney Medical Archives of The Johns Hopkins Medical Institutions.
Reed Pyeritz, a medical geneticist and former student of McKusick’s, has described the impetus for maps depicting the ‘morbid anatomy’ of the human genome to me in this way,
When people started laying out the 23 sets of chromosomes, the ideograms, and then had next to [each] where a gene had been identified, [McKusick] said, ‘that’s all well and good, but you can often map a phenotype [like Marfan syndrome] to a specific site on a chromosome before you know what the cause is’.Footnote 69
In effect, the ‘morbid anatomy’ diagrams acted to breakdown the visual divide between laboratory and clinical knowledge: the anatomical markers of clinical disorders could also be located and observed within the human genome, at the visual level of chromosomes. As McKusick saw it (according to Pyeritz), ‘this is what a geneticist does’.Footnote 70 Just as Morgagni had associated clinical disorders with the anatomy of particular organs during the 18th century, McKusick felt that a major goal of 20th century geneticists should be to give diseases a new, neo-Vesalian basis by locating them in discrete, visible regions within the human genome.
Updated versions of McKusick’s ‘morbid anatomy’ of the human genome appeared frequently in print throughout the 1980s. For instance, morbid anatomy maps were printed in the 1983 and 1986 editions of Mendelian Inheritance in Man, McKusick’s well-known and longstanding catalogue of genetic diseases. New editions of the morbid anatomy of the human genome were also included in various medical genetics texts during the 1980s including, Genetics in Medicine (1986) and The Metabolic Basis of Inherited Disease (1989). In addition, the morbid anatomy was published along with an interview of McKusick in a 1984 issue of the Journal of the American Medical Association, and in a four part series in the journal Medicine between 1986 and 1988.Footnote 71 Indeed, during the mid-1980s, this ‘neo-Vesalian’ depiction of the human genome had made its way more widely into the mainstream medical literature.
With each morbid anatomy figure, McKusick captured the ongoing ‘dissection’ of the human genome, a term that he often used in order to acknowledge the contributions of new molecular techniques in gene and disease mapping. The introduction of restriction enzyme analysis in the 1970s offered geneticists with a ‘scalpel’, as McKusick put it, which could be used to isolate and examine genes at the molecular level.Footnote 72 While geneticists could not observe genes in the same way that they could chromosomes, visual analysis still played a prominent role in molecular genetics research. After taking advantage of restriction enzymes to cut out specific portions of DNA from the genome, geneticists used gel electrophoresis and radiological tags to separate out and visually distinguish genetic variations at the molecular level. While this sort of research remained in the realm of PhD geneticists, McKusick purposely made metaphorical references to using a ‘scalpel’ to ‘dissect’ the ‘morbid anatomy’ of the human genome in order to demonstrate that the genome was as much a relevant object of study for clinically oriented MD medical geneticists, as it was for PhD trained human and molecular geneticists.
8. The Human Genome Goes Full Circle
In this paper, I have demonstrated how standardised depictions of the human chromosomes evolved during the 1970s and 1980s, just as conceptions of the human genome were also shifting. Between 1971 and 1981, chromosomal ideograms became increasingly linearised and densely packed with almost 1000 distinct bands, providing a standardised visual language for dividing up and identifying discrete locations on each human chromosome. At the same time, beginning around 1980, various clinically oriented geneticists, including Victor McKusick and Charles Scriver, began visually representing and alluding to the human genome in anatomical terms. This way of thinking about the genome, as both observable and embodied, led to a noticeable shift in how the term ‘genome’ was used and defined in medical genetics and general medical textbooks between the 1970s to the mid-1980s. No longer referred to abstractly as ‘all of the human genes’, definitions and representations of the human genome became increasingly embedded in, and bounded by, visual representations of the human chromosomes.
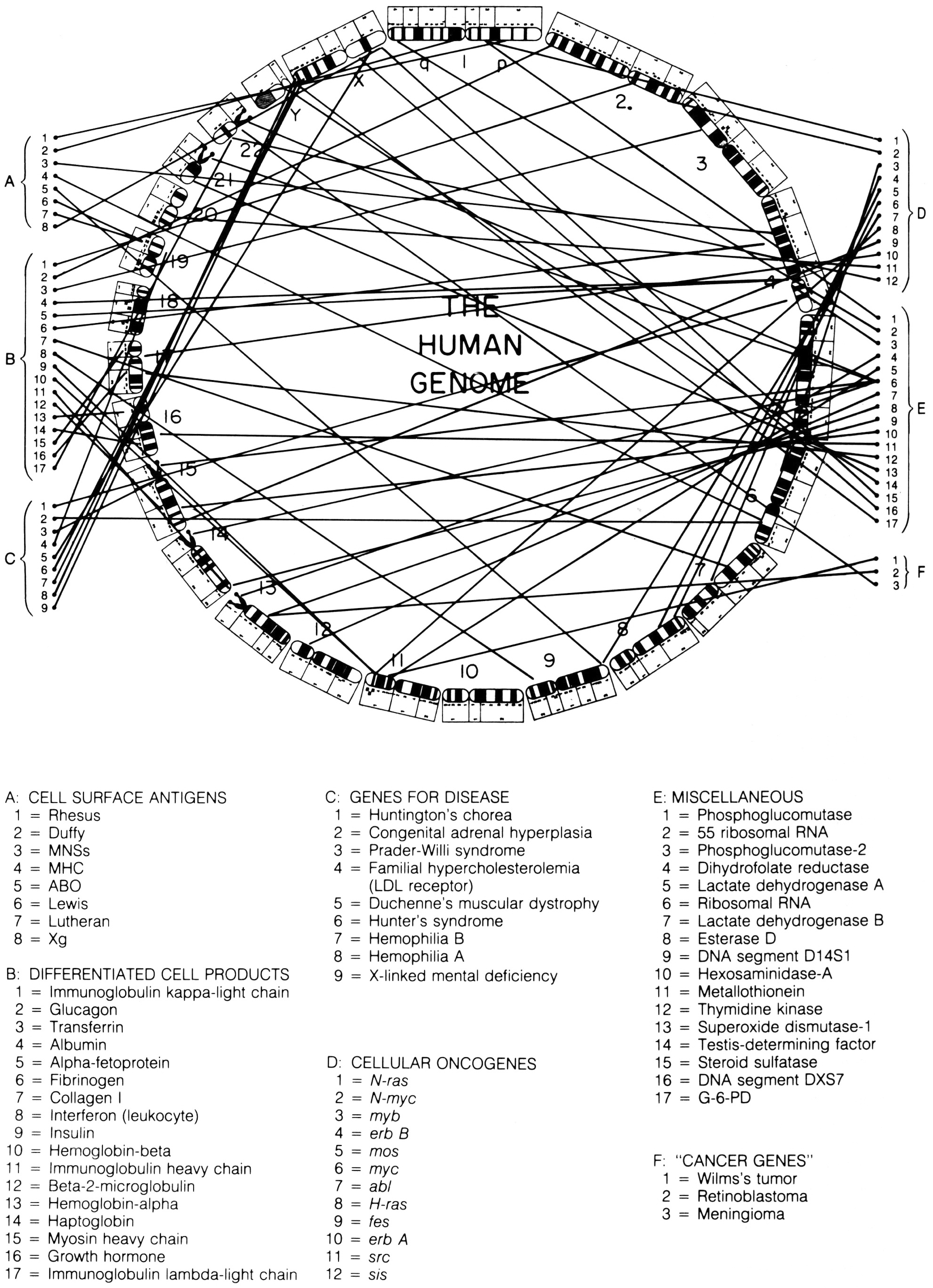
In the 1987 edition of Harrison’s Principles of Internal Medicine, a new figure was added to the text, which perfectly captures the simultaneous evolution of standardised chromosomal ideograms and anatomical representations of the human genome. This image combines selected elements of McKusick’s human gene map and ‘morbid anatomy’ of the human genome, but in a new way. Previously, the ideograms in these figures had been organised as they would be in a standard karyotype: with chromosomes lined up side by side, and in order by size. In this figure, however, chromosomal ideograms were lined up end-to-end, from chromosome 1 to 22, then Y and X, in a circle with ‘THE HUMAN GENOME’ printed in the middle (figure 5).Footnote 73

Figure 5: Circular depiction of chromosomal ideograms, arranged end-to-end and making up the entire human genome. From Braunwald et al. Harrison’s Principles of Internal Medicine, Eleventh Edition (1987). Reprinted with permission from McGraw-Hill Companies, Inc.
This way of depicting the human genome is interesting for a number of reasons. First, and most importantly for this paper, it takes the ongoing process of linearising the chromosomal ideograms, thus making them more ‘genomic’, one step further: the human genome was now depicted as a continuous linear arrangement of all 24 distinct human chromosomes, placed end-to-end. While each chromosome remains physically distinct, this image offers a particular view of the genome, allowing it to be seen and mapped as a continuous whole, instead of in 24 parts.
Another fascinating result of this way of illustrating the human genome is how similarly it is in setup to the standard depiction of bacterial and viral genomes. Instead of being broken down into chromosomes, bacterial genomes are composed of one undivided loop of genetic material. Such genomes can be sequenced or mapped continuously, beginning and ending at any point. Depicting the human genome, which is physically divided into many pieces, in this way suggests a similar conceptual continuity. The human genome becomes a single, bounded entity that can be broken down visually into a continuous series of systematically named chromosomal bands, instead of into 24 chromosomes, each with their own individual unique banding pattern.
Indeed, ‘THE HUMAN GENOME’ captures in one image the various conceptual shifts that I trace in this paper. In being physically embodied by the human chromosome set, the genome was presented as a component of the human anatomy. As part of becoming embedded in the chromosomes, however, the presumed linear arrangement of its genes was not lost. Rather, as I have described, presumptions of linearity could be seen in decisions about how standardised chromosomal ideograms were drawn. The arrangement of these chromosomal ideograms from end-to-end into a closed circle demonstrates dual understandings: the human genome was at once physically embodied by discrete entities, and yet conceptually continuous and linear. In this image, therefore, we see the human genome presented as a discrete scientific object: one that could be understood, described and visually observed as a physically bounded whole.
9. Conclusion
During the 1980s, influential medical geneticists actively began promoting the idea that genetic mapping was more than a cartographic exercise, it was also a distinctly anatomical project. In the titles of multiple papers, published in both genetics and medical journals, Victor McKusick suggested that, going forward, the human genome would not just be seen ‘through the eyes of Mercator’, but also as part of the Vesalian tradition, which would be familiar to the MD trained clinician.Footnote 74 Indeed, throughout the 1980s, McKusick and his colleagues in medical genetics sought to reframe the human genome as being more than an abstract, informational concept. The genome, they argued, was an observable, physical and dissectable part of the human anatomy, characteristics that placed it fully in the realm of traditional medical practice.
This transition from the intangible to the embodied human genome was made possible by a number of concurrent developments. Of major significance were improvements, going back to the early 1950s, in the observational analysis of human chromosomes, which made these entities increasingly visible, distinguishable and standardisable. Along with this came international efforts to improve the representation of chromosomes as idealised ideograms. With the introduction of new chromosomal banding techniques during the 1970s, a set of banded ideograms was produced, and a corresponding visual nomenclature was developed. This naming system came to be used by clinicians and geneticists alike for identifying discrete chromosomal ‘addresses’.Footnote 75 As part of this, researchers involved in gene mapping took up these standardised coordinates for use in identifying the physical locations of genetic traits along the chromosomes.
As genetic traits became associated with physical addresses on each human chromosome, researchers increasingly began to visually represent genes, and indeed the entire genome, as being embodied by the chromosomes, and thus a physical part of the human anatomy. McKusick in particular pushed to promote this anatomic metaphor, publishing eight papers in scientific and medical journals during the 1980s that directly made this connection. In this pursuit, McKusick was aided by fellow medical geneticist Charles Scriver, who linked genomic gene and disease mapping to the Vesalian tradition of medicine in his scientific publication and conversations with leading funders of medical research. Between Scriver’s ‘neo-Vesalian’ references and McKusick’s ‘Morbid Anatomy of the Human Genome’ diagrams, the concept of the human genome as a medical entity was fully in place by the mid-1980s.
These moves, towards making the human genome an object of medical analysis, I argue, were representative of the broader rise of postwar biomedicine. In his ‘morbid anatomy’ figures, McKusick used standardised representations of biologically normal human chromosomes, in order to depict the location of suspected genetic pathologies, which caused disease. In doing so, McKusick took a set of standardised representations from the biological study of human genetics, and used them as a basis for depicting knowledge from the medical area of pathology. This melding of the normal and the pathological in the same physical and conceptual space has been central to the development of postwar biomedicine. Bringing together the historically distinct traditions, languages, and institutions of biology and pathology has involved the creation of new, hybrid locations and ‘platforms’ for doing biomedicine, as well as novel languages and objects of study.Footnote 76 Medical genetics, therefore, can be seen as a microcosm of this ongoing postwar biomedical hybridisation, with the framing of the human genome as a relevant object of study for both MD clinicians and PhD geneticists, being reflective of this process in action.
In order to fully grasp the development of postwar biomedicine, historians and sociologists of science and medicine have looked to the building of new spaces, languages, professions and institutions.Footnote 77 As part of this, I argue that scholars should also look to the observational practices of postwar biomedicine, and through these approaches, at the formulation of common objects of study among practitioners in newly developing interdisciplinary areas. In the case of medical genetics, a common object and language were created, based on chromosomal observation, at the same time that influential clinicians were actively promoting the genome as being part of the human anatomy, and thus part of traditional medical practice. Further study of the human genome, and its important place in the building of contemporary biomedicine will continue to elucidate this ongoing process in which studies of the normal and the pathological have been brought together professionally, institutionally and conceptually.