Introduction
Ruminants harbour symbiotic microbes in their gastrointestinal tract that are essential for the host animal development and adaptation to the environment. Among the different groups of microbes inhabiting the ruminant’s forestomach, bacteria are the most diverse and play an indispensable role in health and nutrition. In the rumen, three different subpopulations of bacteria can be distinguished based on their localisation: (i) a planktonic population composed of bacteria free in the rumen fluid; (ii) a population attached to feed particles; and (iii) a population attached to the rumen epithelium (Cheng and Costerton, Reference Cheng and Costerton1986). The latter is also known as the bacterial epimural community (BEC; Mead and Jones, Reference Mead and Jones1981). This community has been less studied than the others, probably because it represents less than 1% of the total ruminal microbial biomass (Czerkawski, Reference Czerkawski1986) and its contribution to the rumen capacity to ferment feeds is relatively minor. However, the BEC plays an important role in the hydrolysis of systemic urea that diffuses from the blood across the rumen wall (Wallace et al., Reference Wallace, Cheng, Dinsdale and Ørskov1979) and, consequently, on nitrogen metabolism. Tissue recycling and oxygen scavenging are other functions that have also been attributed to BEC (Cheng et al., Reference Cheng, McCowan and Costerton1979).
The BEC was mainly described in the 1970s and 1980s using microscopy and cultural techniques (Bauchop et al., Reference Bauchop, Clarke and Newhook1975; Wallace et al., Reference Wallace, Cheng, Dinsdale and Ørskov1979; McCowan et al., Reference McCowan, Cheng and Costerton1980; Dehority and Grubb, Reference Dehority and Grubb1981). These phenotype-based techniques underestimate biodiversity since they do not take into account the uncultured bacteria and do not discriminate between genetically close species. This perhaps offers an explanation for the contradictory results reported in some earlier studies describing BEC as either specific to the rumen epithelium (McCowan et al., Reference McCowan, Cheng and Costerton1980) or similar to that found in rumen contents (Dehority and Grubb, Reference Dehority and Grubb1981; Mead and Jones, Reference Mead and Jones1981). The advent of genetic techniques has revealed an extensive microbial diversity that was previously undetected with culture-dependent methods (Stahl et al., Reference Stahl, Flesher, Mansfield and Montgomery1988; Pace, Reference Pace1997). Indeed, recent work using cloning and sequencing has shown both the diversity of the BEC and its differences with the population present in rumen contents (Cho et al., Reference Cho, Cho, Shin, Lim, Hong, Choi, Kang, Lee, Kim, Kim and Yun2006). The clone library approach, preferred by some researchers as it allows the genotypic identification of bacterial species, is, however, fastidious and not adapted for monitoring temporal and/or diet-induced changes in the rumen microbial population when groups of animals are used. Fingerprint profiles analyses based on 16S rDNA sequences are currently the techniques more adapted to study bacterial communities. In addition to their relative ease of implementation and the ability to compare several samples, these techniques are promoted as better estimators of biodiversity (Pedros-Alio, Reference Pedros-Alio2006).
Diet is a major factor influencing the structure and function of the rumen microbial population. The nature of feeds, as well as the physico-chemical changes induced by their fermentation, is known to favour the development of certain functional microbial ecotypes in the rumen solid and liquid phases (Martin et al., Reference Martin, Fonty and Michalet-Doreau2002). However, it is not well known whether the same types of changes are also true for BEC. In this work, we tested the effect of diet (forage v. concentrate) on the structure of the bacterial population attached to the rumen epithelium using a genetic fingerprinting technique, 16S rDNA polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE). The effect of localisation within the rumen was evaluated at the same time, with comparison of the BEC to the bacterial communities present in the solid and liquid phases of rumen contents.
Material and methods
Animals and feeds
Eight 5-month-old lambs, four males and four females, of similar genetic composition (stock INRA 401) were used in this study. The lambs were allotted into two homogenous groups, based on body weight and sex, which were fed forage (F, 100% alfalfa hay) or a high concentrate (HC, 80% cracked wheat and 20% alfalfa hay on a dry-matter basis) diet. Feeds were offered once daily at 0800 h for ad libitum intake. Animals were housed in individual pens with free access to fresh water and a mineral salt mix. At the start of the experiment, average body weight was 29.5 ± 2.8 kg and 29.5 ± 2.4 kg for the F and HC groups, respectively. Animals were slaughtered at a similar body weight of 38.8 ± 1.3 kg and 39.8 ± 1.3 kg after 5 and 4 weeks of feeding the F or HC diet, respectively. Animals were cared for in accordance with the guidelines for animal research of the French Ministry of Agriculture (Anonymous, 1988).
Sampling of rumen epithelium and rumen content
Lambs were slaughtered at INRA-Theix’s experimental abattoir. The entire gastrointestinal tract was removed immediately after slaughter and samples from the epithelium were taken from five reticulorumen locations considered representative of the bacterial diversity present on the rumen wall (Mead and Jones, Reference Mead and Jones1981). These sites were the roof of the dorsal sac at the point where the cranio–caudal and left–right axes meet (DS1), the roof of the dorsal sac at a caudal position (DS2), the floor of the caudal sac (CS), the floor of the ventral sac (VS), and the right lateral area (LA) at the point where the cranio–caudal and dorso–ventral axes meet. Samples were taken with the help of a biopsy punch (diameter 8 mm), washed with sterile phosphate-buffered saline (PBS) 0.01 mol/l, pH 6.8, snap-frozen in liquid nitrogen and stored at −80°C until processing. The time elapsed between slaughter and collection of samples was less than 10 min in order to minimise post-mortem changes in the tissue and related possible changes in the BEC (Bauchop et al., Reference Bauchop, Clarke and Newhook1975; Mead and Jones, Reference Mead and Jones1981).
In addition to rumen epithelium, the liquid (LP) and solid (SP) rumen content phases were also sampled. The removed reticulorumens were cut open with the help of a sterile scalpel and contents were thoroughly mixed with an alcohol-sterilised spatula. Approximately 300 g of mixed rumen contents were filtered through a polyester monofilament fabric (250-μm mesh aperture). One-ml aliquots of the filtrate (LP) were dispensed in microtubes, centrifuged (10 000 × g, 10 min, 4°C), the supernatant removed and the pellet was stored at −80°C. The retentate (SP) was washed with 50 ml of PBS, filtered as above and stored at −80°C.
DNA extraction and PCR amplification
Total DNA was extracted from samples as described by Yu and Morrison (Reference Yu and Morrison2004). A method specifically developed for the extraction of bacterial DNA from gastrointestinal tissues was also tested for the epithelial samples (Roussel et al., Reference Roussel, Wilks, Harris, Mein and Tabaqchali2005). This method gave similar results in terms of yield and DGGE fingerprint profiles to those obtained with the method of Yu and Morrison (Reference Yu and Morrison2004), developed for gastrointestinal contents, and thus all samples were extracted using this latter procedure.
The V3 variable region of the 16S rDNA gene of bacteria was amplified by PCR with primers 341f- (5′-CCTACGGGAGGCAGCAG-3′) and 534r (5′-ATTACCGCGGCTGCTGG-3′), the forward primer had a GC clamp at its 5′ end (Muyzer et al., Reference Muyzer, De Waal and Uitterlinden1993). The PCR mixture (50 μl) contained 1 × PCR buffer, 1.5 mmol/l MgCl2, 200 μmol/l of each dNTP, 0.25 μmol/l of each primer and 2.5 U HotStartTaq DNA Polymerase (Qiagen, Hilden, Germany). Touchdown PCR was performed with an initial denaturation step of 95°C for 15 min; followed by 10 touchdown cycles of 94°C for 30 s, 61°C (−0.5°C per cycle) for 30 s and 72°C for 1 min; followed by 25 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 1 min; and a final elongation step of 72°C for 30 min to eliminate artifactual double bands (Janse et al., Reference Janse, Bok and Zwart2004).
All PCR products were analysed by electrophoresis on 2% agarose gels containing ethidium bromide to check their size and estimate their concentration using a low DNA mass ladder (Invitrogen, Carlsbad, CA, USA) and an imaging system (Chemimager; Alpha Innotech, San Leandro, CA, USA).
Denaturing gradient gel electrophoresis
Polyacrylamide gel at an 8% concentration was prepared with a denaturant gradient between 40% and 60% (urea formamide). One hundred percent denaturant was defined as 7 mol/l urea and 40% (v/v) formamide (Muyzer et al., Reference Muyzer, De Waal and Uitterlinden1993). Approximately 100 ng of PCR product were applied per well. Gel was submerged in 0.5 × TAE (Tris-Acetate-EDTA) buffer (40 mmol/l Tris base, 40 mmol/l glacial acid acetic, 1 mmol/l EDTA) and electrophoresed for 5 h at 60°C using a fixed voltage of 200 mV in a DGGE-2001 (CBS Scientific Co., Solana Beach, CA, USA). Gels were silver stained using a commercial kit (Bio-Rad Laboratories, Hercules, CA, USA). Due to the large number of samples, several gels were required to carry out the analysis. In order to normalise for differences between gels, a PCR product from a rumen sample was used on every gel (two lanes per gel) and the profile of this was used as a standard during gel normalisation and analysis. The percentage of similarity between gels’ standards was 77.5 ± 10.6 (mean ± s.d.).
DGGE gel analysis
Gel images were acquired using an optical density calibrated scanner (ImageScanner; GE Healthcare, Piscataway, NJ, USA) at a spatial resolution of 400 d.p.i. and each band was considered an operational taxonomical unit (OTU). Images were analysed using ImageQuant TL software (GE Healthcare) and GelCompar II version 4.0 package (Applied Maths, Kortrijk, Belgium). ImageQuant TL was used to quantify the banding profiles within each profile by determining the total number of bands (S), the peak surface of each band (ni) and the sum of all the peak surfaces of all bands (N) (Fromin et al., Reference Fromin, Hamelin, Tarnawski, Roesti, Jourdain-Miserez, Forestier, Teyssier-Cuvelle, Gillet, Aragno and Rossi2002). This information was used to calculate the community biodiversity using three indices: (i) the Shannon index (H) calculated with the formula H = −Σ(ni/N) ln (ni/N); (ii) the dominance index (c) calculated with the formula c = Σ(ni/N)2; and (iii) the evenness index (e) calculated with the formula e = H/ln S (Odum, Reference Odum1971). These indices were then processed by analysis of variance using the MIXED procedure of Statistical Analysis Systems Institute (SAS, 1999). The statistical model included animal, diet, localisation (corresponding to the sampling sites for the rumen epithelium and the nature of the phase for the rumen content) and diet × localisation interaction. Animal within diet was considered as random effect. Because sampling site was not significant for epithelial samples, the data were grouped and compared with the LP and SP of the rumen contents using the same model stated above. Effects were declared significant at P < 0.05.
GelCompar II was used to normalise and compare all the DGGE profiles using hierarchical clustering to join similar profiles into groups (Fromin et al., Reference Fromin, Hamelin, Tarnawski, Roesti, Jourdain-Miserez, Forestier, Teyssier-Cuvelle, Gillet, Aragno and Rossi2002). To this end, all the images of DGGE gels were matched using the internal control sample and the bands were quantified after a local background subtraction. A tolerance in the band position of 1% was applied. The similarity among profiles was calculated with the Pearson product–moment correlation coefficient, recommended for the analysis of complex profiles (Savelkoul et al., Reference Savelkoul, Aarts, de Haas, Dijkshoorn, Duim, Otsen, Rademaker, Schouls and Lenstra1999), and the clustering was done with the unweighted pair-group method using arithmetic averages (UPGMA). A significance test based on pairwise similarity measures was used to compare the community profiles of different groups of samples (Kropf et al., Reference Kropf, Heuer, Gruning and Smalla2004). This permutation test was done using the PROC IML procedure of SAS (1999) with 105 random permutations and based on the model of Kropf et al. (Reference Kropf, Heuer, Gruning and Smalla2004), which allows the utilisation of more than one gel in the analysis. An additional permutation analysis was also done using the control samples, grouped by gel, to further test whether the effect of gel was significant.
Results
Bacterial diversity of rumen epithelial and rumen content samples
The biodiversity of BEC, as assessed by the indices of Shannon, evenness and dominance calculated from the DGGE profiles, was not influenced by sampling site or diet (data not shown). There was a non-significant trend for samples taken from the upper part of the rumen to have higher values for the Shannon index than samples taken from the lower part (2.827 v. 2.531 for DS1 and VS, respectively, P = 0.16). The comparison of BEC with the bacterial population of rumen content samples showed differences (P < 0.01) for all three indices (Table 1). In contrast, there was no effect of diet or diet × bacterial population interaction on these biodiversity indicators. The bacterial population attached to the rumen epithelium had mean values for the index of Shannon and dominance that were halfway of those for the SP and LP of rumen contents. The SP had the highest value for the Shannon index and the lowest for the index of dominance.
Table 1 Biodiversity indices from DGGE fingerprints of the rumen epimural, liquid- and solid-associated bacterial communities
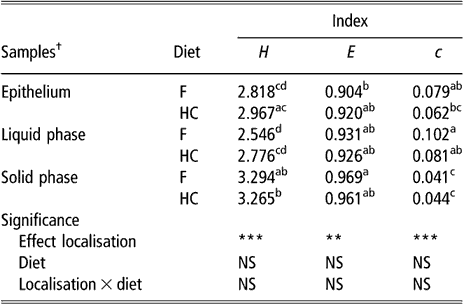
DGGE = denaturing gradient gel electrophoresis, NS = not significant, F = forage diet, HC = high-concentrate diet, H = Shannon’s index, e = evenness, c = dominance.
a,b,c,dWithin a column, means with different letters are significantly different (P < 0.05). NS: P > 0.05; **P < 0.01; ***P < 0.001.
†Means of eight lambs for liquid and solid phases (s.e. = 0.1522, 0.0248 and 0.0126 for H, e and c, respectively). Means of eight lambs and five sampling sites per animal for epithelial samples (s.e. = 0.0979, 0.0184 and 0.0073 for H, e and c, respectively).
Effect of localisation on the bacterial community structure
Samples were allocated to four DGGE gels. A permutation test that included in the model all electrophoretic runs was used for the comparison of groups of samples. In addition, a permutation test analysis that considered standard samples from each gel as a group indicated that differences due to electrophoretic run were not significant (P > 0.05).
Similar to the results obtained using biodiversity indices, cluster analysis of BEC samples obtained from DGGE profiles indicated no differences in BEC structure associated with sampling site or diet (data not shown). In subsequent analysis, the two sites most different in terms of anatomical localisation, e.g. VS and DS1, were selected as representatives of the rumen epithelium. When the clustering analysis was applied to rumen content and the selected epithelial samples, the dendrogram topology revealed that epithelial samples were distinctly grouped from the SP and LP samples of rumen contents (Figure 1). The difference between sites is illustrated in Figure 2 using samples from one representative animal fed either F or HC diets as an example. The dendrogram obtained had two main nodes. One of them grouped the rumen content samples of F-fed animals. This node was further branched at a second level that separated the LP from the SP samples. The other node, which contained the rest of the samples, was subdivided into two groups containing the rumen epithelial samples on one side and the rumen content samples of animals fed the HC diet on the other. Permutation test analysis of this data set confirmed that BEC consistently differed from the bacterial community associated with the LP and SP samples (P < 0.01; Table 2) and that the BEC was unaffected by diet, unlike the LP and SP bacterial communities (P < 0.05; Table 2). The LP and SP samples of F-fed animals were also significantly different (P < 0.05). In agreement with the cluster analysis, no distinction was observed in the LP and SP sampled from the HC diet (P > 0.05).

Figure 1 UPGMA (unweighted pair-group method using arithmetic averages) dendrogram generated from bacterial denaturing gradient gel electrophoresis (DGGE) profiles. Samples are from the rumen epithelium ventral and dorsal sacs (VS and DS1) and the rumen liquid (LP) and solid (SP) phases, taken from eight lambs that received a forage (F) or a high-concentrate (HC) diet.

Figure 2 Typical denaturing gradient gel electrophoresis (DGGE) profiles of the bacterial community attached to the rumen epithelium (ventral and dorsal sacs, VS and DS1, respectively) and associated to the rumen liquid and solid phases. Samples taken as example are from animals F and D fed a forage (F)-based and a high-concentrate (HC)-based diet, respectively.
Table 2 Results of permutation test comparing bacterial DGGE fingerprints of the rumen epimural, liquid-, and solid-associated bacterial communities†
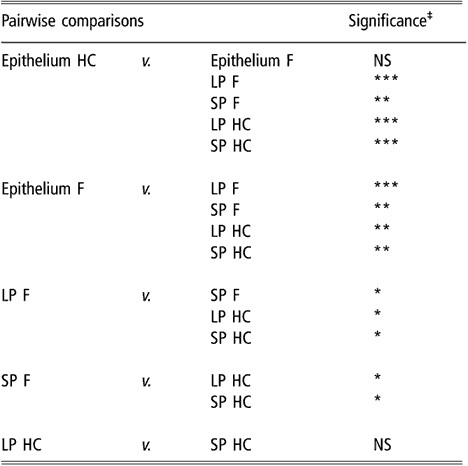
DGGE = denaturing gradient gel electrophoresis, HC = high-concentrate diet, F = forage diet, NS = not significant, LP = liquid phase, SP = solid phase.
†Samples are from the rumen epithelium (five sampling sites), LP and SP from eight lambs that received a F diet or a HC diet.
‡NS: P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
Bacterial diversity plays a central role in the functioning and productivity of microbial ecosystems (Cardinale et al., Reference Cardinale, Palmer and Collins2002; Bell et al., Reference Bell, Newman, Silverman, Turner and Lilley2005). Diversity can be studied by different methods. The construction of clone libraries is a frequently used method but it is less efficient in retrieving the diversity present in complex ecosystems (Pedros-Alio, Reference Pedros-Alio2006). In this work, we chose the fingerprinting technique PCR-DGGE, to advance our understanding of some of the parameters that govern the structure of the rumen epimural community and how it is affected by the environment, e.g. diet. DGGE analysis clearly showed that the bacterial community attached to the rumen epithelium was (i) different from LP and SP bacterial communities present in the rumen and (ii) that its structure was not affected by diet or sampling site. This distinction between the bacterial population attached to the rumen wall and those in rumen content is opposed to previous reports that described these populations as taxonomically and physiologically similar (Dehority and Grubb, Reference Dehority and Grubb1981; Mead and Jones, Reference Mead and Jones1981). Other authors using similar culture-based techniques, however, described a unique tissue-adherent bacterial population in the bovine rumen (Cheng et al., Reference Cheng, McCowan and Costerton1979). More recent works based on 16S rDNA gene sequences of clone libraries also support the uniqueness of this population (Mitsumori et al., Reference Mitsumori, Ajisaka, Tajima, Kajikawa and Kurihara2002; Cho et al., Reference Cho, Cho, Shin, Lim, Hong, Choi, Kang, Lee, Kim, Kim and Yun2006). Permutation analysis indicated significant differences among samples from the epithelium, rumen contents from HC-fed animals and rumen contents from F-fed animals, even though the similarity in values between the clustered profiles in the dendrogram were not that high (Figure 1). This low within-group similarity could be due to animal differences, despite their similar genetic composition and common rearing environment before and during the study. Friswell et al. (Reference Friswell, Gilbert, Allison, Stratford, Telfer and Mcbain2006) reported a higher degree of similarity between the faecal flora of individual rats of the same strain. Laboratory rat strains are certainly more genetically homogeneous than the sheep stock used in this study, and intrinsic differences in the gastrointestinal population sampled, e.g. rumen v. faeces, may have contributed to the dissimilar results. Other studies in ruminant species also reported large animal differences in the diversity of the bacterial population present in rumen contents (Edwards et al., Reference Edwards, Bequette, McKain, McEwan and Wallace2005; Larue et al., Reference Larue, Yu, Parisi, Egan and Morrison2005). Due to the intimate contact between animal tissue and BEC, the host influence on this population is stronger than for bacteria in rumen contents (Cheng and McAllister, Reference Cheng and McAllister1997). The absence of differences detected in BEC due to dietary composition may also be explained by the same reason. However, it should be noted that subtle variations in the community might be undetected using this technique. DGGE has an abundance limit of 1% (Fromin et al., Reference Fromin, Hamelin, Tarnawski, Roesti, Jourdain-Miserez, Forestier, Teyssier-Cuvelle, Gillet, Aragno and Rossi2002), and thus the contribution to diversity of less-abundant taxons is underestimated. Nevertheless, these rare taxons have been proposed to be less relevant for the functioning of bacterial ecosystems (Pedros-Alio, Reference Pedros-Alio2006). In addition to the distinct structure of BEC, DGGE analysis differentiated the LP bacterial population from that of SP in F-fed lambs. This could be explained by the increased proportion of fibre-degrading bacteria associated with solids reported for this type of diet (Michalet-Doreau et al., Reference Michalet-Doreau, Fernandez, Peyron, Millet and Fonty2001). For the concentrate-rich diet, the absence of differentiation between the liquid- and solid-associated bacteria is probably due to a large amount of small starch granules in LP that makes substrate separation between phases less defined.
The higher diversity of the SP population correlates with the high number of microbes found in close association with solids in the rumen (Cheng and McAllister, Reference Cheng and McAllister1997). Shannon values also suggest that BEC, despite being less important in terms of biomass, was more diverse than the LP population. BEC, situated at the interface between the host tissues and rumen contents, is in contact with a variety of substrates and other microscale conditions. This heterogeneous environment could promote bacterial diversity as shown in other ecosystems (Horner-Devine et al., Reference Horner-Devine, Lage, Hughes and Bohannan2004). The biodiversity measured with DGGE should be interpreted with caution though, as there is a limit in the number of OTU (bands) that can be visualised in a gel (Loisel et al., Reference Loisel, Harmand, Zemb, Latrille, Lobry, Delgenes and Godon2006).
In conclusion, using PCR-DGGE, we have shown that BEC was different from the communities present in rumen contents of lambs. The bacterial population attached to the rumen epithelium was not affected by diet, indicating that the host animal may have a strong influence on the BEC structure. Similarity of BEC between animals was low, whether this difference in similarity has any influence on the function of this community remains to be determined.
Acknowledgements
S. Sadet is the recipient of an INRA-Auvergne region fellowship. The authors are grateful to J. Teuma, M. Bernard and P. Faure for the care of animals, the personnel of INRA-Theix’s experimental abattoir, D. Graviou and Y. Rochette for their technical assistance, F. Glasser for statistical advice, and P. Nozière for critical reading of the manuscript.





